Mbirn: DTI Calibration Studies
From NAMIC Wiki
Home < Mbirn: DTI Calibration Studies
GOALS
- To study effect of various imaging parameters on DTI results. In the initial phase of the study, we will concentrate on influence of SNR, b-value, gradient orientation, and echo time on diffusion anisotropy measure.
- Plans for Jun 2005 - Oct 2005 (S. Mori)
- Acquisition of working datasets
- Dataset #3: Multiple b-values to study effects of b-values: May - June / 2005
- Dataset #4: Multiple echo time to study effects of echo time: July / 2005
- Data analysis I: Effect of SNR on anisotropy
- Data acquisition and processing are done. We'll analyze the data to study the SNR-anisotropy relationship: May - June / 2005.
- I hope that the current data are enough for publication but we never know until we finish the analysis. Depending on the results, we may need additional data. Manuscript should be ready by October / 2005.
- Data analysis II: Effect of gradient orientations
- Data acquisition and processing are done. Initial data analysis is going well, but we need more time for data analyses. We'll have a good idea about the results in May / 2005.
- I hope this result will be publishable too. We'll see.
- Acquisition of working datasets
DTI VISUALIZATION UPDATES
DTI CALIBRATION UPDATES
- May 17, 2005 (S. Mori)
- Acquisition of working datasets
- We finished acquisition of two types of datasets (Dataset #1 and Dataset #2), each repeated 3 times on the same subjects. The 3 repetitions were performed on 3 different days.
- Data will be posted somewhere in the BIRN site.
- Both types of datasets have the following imaging parameters:
- Acquisition of working datasets
B0: 1.5T Philips TE: 100ms B: 1000 s/mm2 SENSE: p=2 TR:longer than 8s Matrix: 96 x 96 (zerofilled to 256x256) FOV: 240 x 240 (2.5 mm resolution) Gradient:Jones 30
Differences are:
Dataset #1:
-Image slice: 50 (whole brain coverage)
-Imaging time: ~4.5 min + 2-3 min reconstruction time
-Repetition: 5 - 6 times (saved separately)
Dataset #2:
-Image slice: 20 (partial brain coverage)
-Imaging time: ~2.5 min + 1-2 min reconstruction time
-Repetition: 10 - 12 times (saved separately)
- ** Data analysis I: Effect of SNR on anisotropy
- Anisotropy maps (FA) were created from single signal average. For example, from the Dataset #1, we can create 5 - 6 FA maps with single average. Similarly, we can create 2 - 3 FA maps with 2 signal average.
- All FA maps with all possible combinations of signal averaging were created.
- After ISMRM, we'll resume the study to analyze relationship between FA values and the number of averaging (SNR).
- We would like to encourage BIRNers to use these datasets to study effects of SNR on other DTI endpoints such as fiber orientations and fiber tracking.
- Data analysis II: Effect of gradient orientations
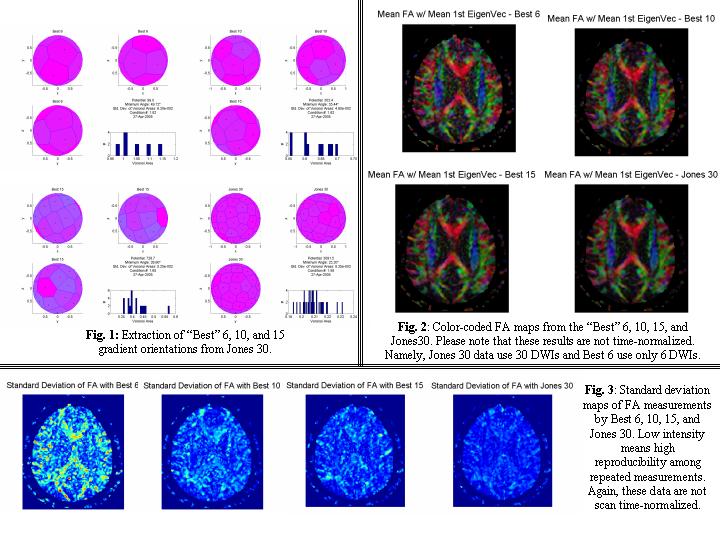
- Following Steve's suggestion, we sub-sampled 6, 10, and 15 gradient tables from Jones 30 orientation.
- We first analyzed quality of the sub-sampled gradient orientations in terms of the uniformity of the gradient angles in 3D space (Fig. 1). Then we extracted subsets of DWIs from the Jones 30 experiments.
- Results are shown in Fig. 2 and Fig. 3 for absolute FA maps and standard deviation maps (reproducibility among repeated measurements).
- Please note that these data are not time-normalized and, for example, comparison between Best 6 and Jones 30 is not fair at this point (Best 6 uses only 6 DWIs vs 30 DWIs for Jones 30). We are currently working on the time-normalized analyses.
- Feb 14, 2005 ( J. Jovicich)
- Susumu, Steve & Jorge met over the phone for updates on Feb 8
- DTI Calibration plans for defining optimum multi-site protocol
- First, find the optimum # of averages (SNR) for FA reproducibility. For this a single subject will be scanned in three separate 1-hour sessions. In each session 12 identical DTI scans will be acquired. Grouping them in different numbers will enable us to look into different averaging schemes. FA reproducibility will be assessed across the three sessions, as a function of # of averages. The hypothesis is that there will be an optimum # of averages above which the reproducibility does not significantly increase. This is the # we are looking for so that we use it for the following tests that optimize the other parameters.
- Second, with the optimum # of averages, and using a 1-hour scan, investigate FA reproducibility effects using a range of b-values.
- All of this will be using SENSE in a 6-channel parallel coil, which inherently gives less distortions (short echo trains) than single-channel RF coils. Allen's distortion correction cannot yet be tested because it needs to be first extended for multi-channel coils, and then ported to the Philips platform.
- For this work Susumu has a 1/2 postdoc that can help with the data acquisition and some of the data processing. Susumo would like to have Boston help with the reproducibility statistical evaluations and it's spatial components. Steve Pieper proposed that BWH could potentially help by doing a similar analysis to what they did with fMRI reproducibility (use of STAPLE method for assessing sensitivity and stability through ROC curves). Jorge will also look into using Silvester's and maybe other MGH help (Doug?).
- Susumu has his scans for the first part of this study scheduled for before the March Meeting. He will try to bring some preliminary analysis to the meeting to facilitate discussions. He will send the mBIRN MRI calibration group any preliminary result slides that can be shared.
- DTI integration with Software & Visualization Tools
- 3D Slicer needs to be extended to provide a more flexible front end for visualizing DTI data. To this end, Steve is interested in having sample of DTI data from currently used protocols at mBIRN & fBIRN sites. Jorge will contact the human BIRN sites to follow up on this request.
- As a starter, Steve can use the the JHU Public DTI database for integration purposes. This database contains 1.5T Philips, MP-RAGE, structural T2 and single-shot EPI, 2.5 mm^3 isotropic voxels, 6-channel parallel imaging, 30 diffision encoded directions, 4-min per acquisition, 3 acquisitions (AC-PC alignment). Due to hardware limitations the images from the 6 channels are averaged on the fly and saved, as opposed to saving all separate channel data. The image data has no header, and missing details can be obtained from Susumu Mori (susumu@mri.jhu.edu). A reason for which dicom format was avoided was the complexity that having a separate file for each slice generates in DTI.
- A goal for the upcoming Morph BIRN Miami meeting would be to characterize the diffusion MRI format variability across mBIRN & fBIRN.