Mbirn: Efforts to Localize lesions (BELL) project
BIRN Efforts to Localize Lesions (BELL) Project
Background:
This project was proposed at the March 2005 Miami Morph BIRN meeting as new application test that could help drive the integration and application of diffusion analyses tools to structural MRI data. The idea is to create probabilistic maps of individual white matter tracts using tractography and to use the coordinate information to answer the following hypothesis; "Can we superimpose the coordinate information on existing MRI to correlate MRI findings (e.g. T2 lesions, fMRI activation maps) with specific white matter tracts?. The initial test would be to use a white matter atlas created by Susumu Mori and register it to part of the MIRIAD data to look for correlations between lesions, damaged white matter tracks and behavioural results"
UPDATES
July 21, 2005
- July 19 San Diego meeting (S. Pieper)
- A group of us got together tuesday morning: Anders, me, Dave Kennedy, Jacapo (UCSD anatomist), Josh (Jacapo's post-doc), Michele, and Cooper. We reviewed the atlas work from Susumu and Dave. Susumu's atlas loads nicely in slicer. The VETSA freesurfer and DTI data also loads nicely and we believe that the tensor frames are all being correctly calculated. Most of the meeting was spent reviewing the technical details.
- Next steps for S. Pieper: test tensor-based registration techniques to map the JHU atlas to the VETSA data. The current slicer technique will be tested first. Within the next year we expect to also have new ITK-based registration that will come out of the work being done by NA-MIC.
- Follow up discussions (J. Jovicich)
- Steve will send James the atlas from Susumu.
- James will identify a few brains with white matter lesions overlapping atlas labels of interest
- Steve will co-register these subjects' scans to the atlas
- Steve/James/Jorge will help acquire calibration data that will be used to ensure that tensor calculations/registrations are correctly derived regardless of vendor's, slice orientation, head orientation.
- James will include test runs in his lesion reproducibility study (4T GE ,1.5T GE, 3T Siemens). He might find a normal volunteer for this too.
- Jorge will look for possibilities to scan Steve at MGH
- Steve will distribute Gordon's information and help ensure that all the variables of interest are considered in the scans.
June 15, 2005 (J. Jovicich)
- Susumu Mori completed a white matter atlas based on diffusion MRI data from 10 subjects. These data is in MNI-ICBM coordinates, and has been made available to Steve and James. An atlas based on 30 subjects will be complete towards the end of July.
- Next steps are to define the details of how this atlas will be used with the MIRIAD data and also to define which subset of the MIRIAD data should be used for a preliminary analyses.
May 17, 2005 (J. Jovicich)
- Probabilistic DTI-based White Matter Label Atlas
- Susumu is building a white matter probabilistic atlas for use in the MIRIAD data. This collaboration was initiated in March-Miami meeting by Steve and Jim.
- The idea is to create probabilistic maps of individual white matter tracts using tractography and to use the coordinate information to answer the following hypothesis; "Can we superimpose the coordinate information on existing MRI to correlate MRI findings (e.g. T2 lesions, fMRI activation maps) with specific white matter tracts?"
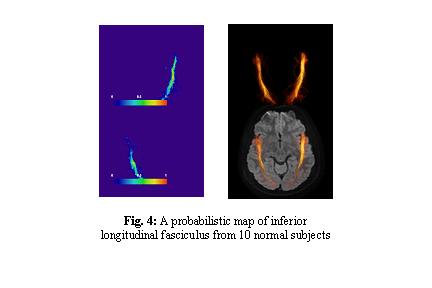
- We finished creation of such probabilistic maps for various white matter tracts (Fig. 4) using data from 10 healthy volunteers. Currently, they are in JHU DTI template coordinates.
- Pending issues are;
- We'll transform the data to ICBM-MNI coordinates to adopt more widely-used coordinates
- We are increasing the number from 10 to 30 volunteers.
- We'll finish these two issues in May and send them to Jorge.
- By mid-June the atlas will consist of 30 averaged subjects and the coordinates will be in ICBM-MRI. This will then be passed on to Steve Pieper for co-registration with the MIRIAD data and then to James MacFall for the lesion/tractography analysis.