Overview
Overview
Delivering on the Promise of the Information Technology Revolution
NA-MIC, the National Alliance for Medical Image Computing, is a multi-institutional, interdisciplinary community of researchers, who share the recognition that modern healthcare demands improved technologies to ease suffering and prolong productive life. Organized under the National Centers for Biomedical Computing six years ago, NA-MIC was created to implement a robust and flexible open-source infrastructure for developing and applying advanced imaging technologies across a range of important biomedical research disciplines. A measure of its success, NA-MIC now is applying this technology to diseases that have immense impact on the duration and quality of life: cancer, heart disease, trauma, and degenerative genetic diseases. The target of this techology is shifting from whole populations to subject-specific analysis. Practicing clinicians and biomedical researchers are intimately aware of gaps in their ability to apply medical knowledge to the needs of individual patients. An abundance of electronic clinical data is produced over the course of an individual's disease progression and treatment, representing an enormous opportunity for improving medical care. The task of interpreting this mass of clinical data, however, has become correspondingly complex, often confounding this opportunity. To borrow a phrase from the intelligence community, the medical community is facing a challenge of "not enough eyeballs per pixel."
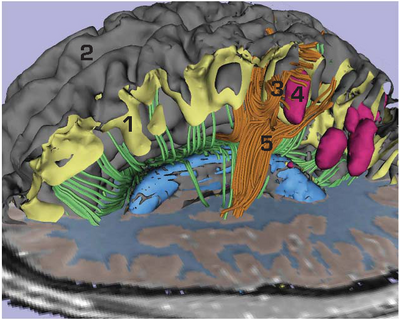
An example of subject-specific analysis on an individual dataset in a patient with moderate traumatic brain injury and intraparenchymal bleeding is shown in the Figure. These results were created using preliminary versions of novel methodologies and tools currently in development by NA-MIC. These methodologies and tools are being developed to aid the clinical assessment of damage and viable treatment options in TBI patients. Analysis of diffusion tensor images allows identification of white matter tracts in the vicinity of one of the lesions. Despite the complexity of the process, this image was constructed on a standard laptop in under 30 minutes. This sort of analysis will eventually enable clinicians to identify issues unique to the patient, guide treatment, and predict outcome in a clinically viable timeframe.
In practice, delivering on the promise of the information technology revolution in biomedical imaging is exceptionally difficult. The barriers to entry include the need to interoperate with scanners and other clinical systems, to organize and present clinical information according to accepted conventions, and to deploy computer systems that can efficiently process data within the time constraints of clinical practice. Since many of these requirements are common across a range of clinical applications, the NA-MIC open source platform allows the most labor intensive development and debugging tasks to be shared by the community for mutual benefit. A well defined, scalable software architecture and rigorous engineering methodology are essential to making software of this scale viable and are hallmarks of the NA-MIC approach.
Impact through Collaboration
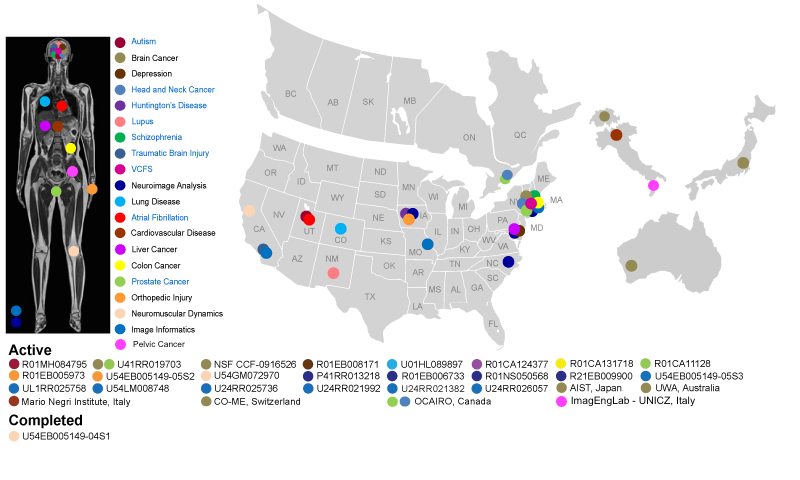
NA-MIC has funded activities at multiple centers in the United States and participated in multiple international collaborations. Each center serves as the nexus for interaction between computer scientists and medical researchers investigating biological problems of vital importance to human health. In the accompanying schematic, each disease process, anatomic location, funding institution, and geographic location has a corresponding color code. Colon cancer, for example, is identified by a yellow dot. Six of these projects are funded by NIH institutes (MH, RR, EB, HL, CA, NS). One additional project is funded by the NSF.
The scope of NA-MIC activities includes both the highly speculative exploration of new mathematical formations of core image analysis techniques and the ongoing effort of delivering and supporting binary distributions of software applications across a range of computing platforms. To address this continuum, the NA-MIC Computer Science Core is organized around two teams: Algorithms and Engineering. These teams bring complementary skills to the technical challenges posed by the driving biologic projects (DBPs). The Algorithms team is led by five senior investigators from four academic institutions. Their combined background provides expertise in variational, statistical, and geometrical approaches to image analysis. The Engineering team is led by five senior investigators from two small businesses, an industrial research facility, and two academic institutions. The joint output of these two teams is the NA-MIC Kit, which embodies a comprehensive set of analysis techniques in a well architected, documented, and widely used platform.
Creating Tools and Infrastructure for Personalized Medicine
NA-MIC is active in two fundamental aspects of biomedical technology: basic research to characterize diseases in populations and clinical decision support to translate this general knowledge to the needs of individual patients. This integration of approaches is referred to as personalized medicine.
The first six years of NA-MIC were focused primarily on basic research in schizophrenia, autism, and lupus, where detailed individual analyses of lesion locations, cortical thickness, white matter architecture, and brain structure morphometry were studied in large populations. Statistics from these group comparisons are essential to defining the range of variation endemic in these conditions. As part of these efforts, NA-MIC invested heavily in the technologies for analysis and visualization of multimodal imaging studies of individuals. Increasingly these tools and approaches have supported translational science in support of patient-specific clinical decision support. Although our computer scientists have strong credentials in this area, NA-MIC's involvement in translational medicine began in earnest with our work in MRI-guided prostate cancer interventions through a funded DBP, in addition to funded collaborations on topics such as CT colonography, radiofrequency liver tumor ablation, and analysis of PET-CT studies of lung tumor characteristics. Our current DBPs reflect the goal of embodying knowledge gained from population analysis in new or modified software systems directed to the benefit of individual patients.
As the current NA-MIC DBPs make concrete technical advances to subject-specific analysis in their respective fields, these advances reflect and inspire important developments in the NA-MIC Kit, which enhance its utility as a research platform and lead to new software solutions. The current DBPs, for example, require improved robustness and efficiency of multimodal nonlinear image registration and segmentation for almost every medical image computing application. All of the DBPs work with multiple or longitudinal image data of individual subjects. These data are used to detect and characterize changes from baseline secondary to disease, trauma, or treatment. This effort requires novel image processing and visualization tools for qualitative and quantitative assessment of serial images. Quantitative evaluation of pathology, a main theme in all four DBPs, necessitates new types of efficient intuitive user-interaction in 2D and 3D displays to efficiently initialize and guide image registration and segmentation procedures. To adapt these tools for clinical use, refinements are needed to render the NA-MIC Kit more compatible with clinical systems, including capabilities for representing clinical data, PACS interfaces, and DICOM networking. Another technical goal common to all NA-MIC DBPs is the delivery of integrated solutions of data, software, and tutorials that represent clinical Best Practices. In this regard, the Computer Science Core is creating or modifying infrastructure for optimizing and validating these Best Practices, while the Service, Training, and Dissemination Cores are designing systems and methodologies for effectively communicating these Best Practices to the wider research community.
To meet the challenges of the information technology revolution, the NA-MIC organization is guided by three principles: (1) Innovation cannot and should not be managed from above, (2) Communication and training across computer science and clinical research disciplines is critical to providing the highest quality biomedical image analysis research, and (3) Open science is good science. These principles are applied in a culture of shared decision-making and mutual responsibility among Core PIs. They have served our Center well in the past and will continue in the future.