Projects:DTIValidation
Back to NA-MIC Collaborations, MIT Algorithms, UNC Algorithms, Irvine DBP 1, Isomics Engineering
DTI Validation
Our Objective is to carry out quantitative and qualitative validation of the DTI tractography tools. These will be applied to a limited set of specific tracts in single data sets and single tractography tools, and on several data sets using at least two tractography programs and by investigators in different laboratories
Description
We have analyzed and validated 11 major tracts in one dataset (Susumu Mori's; 5 subjects) and using one tractography tool (DTI Studio). These were analyzed in several different labs and the quantitative (kappa values) validation was successful and a manuscript is presently in near-final form. We are now analyzing two different datasets (Susumu Mori, Dartmouth) using two tractography tools (DTI Studio, Slicer), quantitative and qualitative comparisons, and two investigators in Fallon's lab. We will also examine these with Guido Gerig's DTI tractography tool.
DTI Studio validation: We have analyzed and validated 11 major tracts in one dataset (Susumu Mori's; 5 subjects) using one tractography tool (DTI Studio). These were analyzed in several different labs. The quantitative (kappa values) validation was successful, and a manuscript has been submitted for publication.
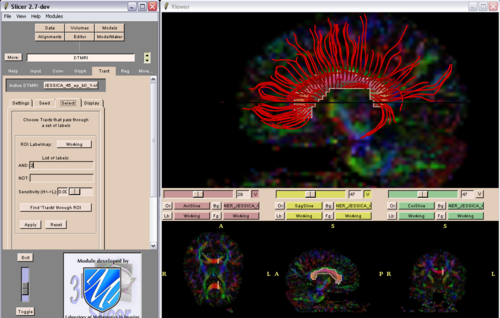
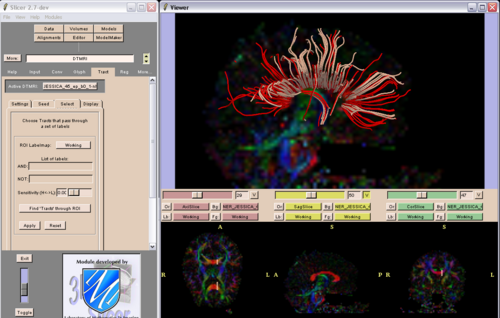
Slicer validation: Due to data format specificity Slicer validation was not feasible on Mori's dataset and the VETSA dataset. On the HUVA dataset Slicer produced a consistent error (abnormal thinness) in generating tracts with a major horizontal component. The HUVA coronal acquisition sequence and slice thickness (5 mmm) are likely contributors to this finding. Slicer produced only very short tracts on the Dartmouth dataset. Our preliminary conclusion is that Slicer is sensitive to the specifics of the diffusion sequence acquisition parameters. Future research is needed to establish critical values for diffusion sequence acquisition parameters that would allow diffusion data processing via Slicer. We solved compatibility issues pertaining to the NRRD format that initially precluded us from using Slicer on UC Irvine collected (Mosaic Siemens 3T Trio) datasets. To asses the Slicer reliability/validity in the same datasets we compared the results of 2 Slicer tracking approaches on the same dataset: a region of interest (ROI) based approach vs a point by point seeding approach. We concluded that the use of the fiber by fiber seeding vs deleting options complicates the creation of standard tracking algorhytms and might in fact results in decreased fiber tracking reliability/validity.
|
|
|
Fig. 1: Corpus callosum fiber differences in the same subject as a result of using different fiber generating algorithms with the point by point seeding function in Slicer 2.7 (DTI Module) |
Key Investigators
- MIT Algorithms: Lauren O'Donnell, Raul San Jose
- UNC Algorithms: Guido Gerig
- Irvine DBP 1: James Fallon, Martina Panzenboeck, Adrian Preda
- Isomics Engineering: Steve Pieper