Projects:RegistrationLibrary:RegLib C03B
From NAMIC Wiki
(Redirected from Projects:RegistrationLibrary:RegLib C03 2)
Home < Projects:RegistrationLibrary:RegLib C03 2v3.6.3
Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
v3.6.3  Slicer Registration Library Case #3B: Diffusion Weighted Image Volume: align with structural reference MRI (ADVANCED/EXTENDED)
Slicer Registration Library Case #3B: Diffusion Weighted Image Volume: align with structural reference MRI (ADVANCED/EXTENDED)
Note that this is the complex/advanced version of this case, which includes preprocessing like resampling the DWI, DTI estimation etc. A simpler version showing only the registration + DTI resampling is here.
Input
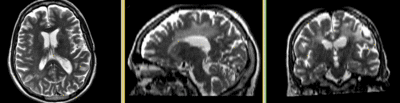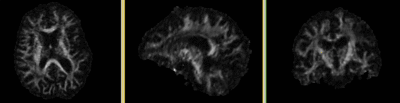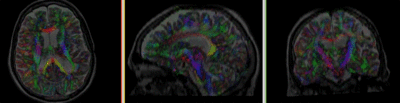
|
|
|
|
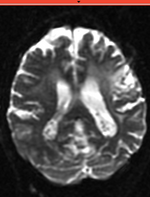
| fixed image/target T2 |
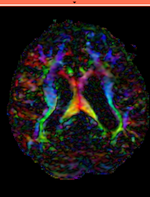
moving image 2a DTI baseline |
moving image 2b DTI tensor |
Modules
- Slicer 3.6.3 recommended modules: BrainsFit, Resample DTI Volume, Diffusion Tensor Estimation
- for the Slicer 3.6.3 version of this case see here
Objective / Background
While this example shares a typical objective of DTI processing, it covers additional issues of a strong initial rotation and strong voxel-anisotropy for the raw DWI image acquired. Goal is to align the DTI image with the structural reference T2 scan that provides accuracte anatomical reference.
Keywords
MRI, brain, head, intra-subject, DTI, DWI
Download
- Image Data:
- RegLib_C03_Data: main registration package: register DTI (Data,Presets, Transforms, zip file 61 MB)
- RegLib_C03_Data_preproc:preprocessing package: resample T2, resample & align DWI, convert to DTI (raw and resampled DWI, zip file 228 MB)
- RegLib_C03_DATA_results: resampled DTI (registered DTI, zip file 39 MB)
- Presets:
- Documentation
Input Data
- reference/fixed : T2w axial, 0.4mm resolution in plane, 3mm slices
- moving: Baseline image of acquired DTI volume, corresponds to T2w MRI , 0.9375 x 0.9375 x 1.4 mm voxel size, oblique
- Tensor data of DTI volume, oblique, same orientation as Baseline image. The result Xform will be applied to this volume. The original DWI has 26 directions, the extracted DTI volume has 9 scalars, i.e. 256 x 256 x 36 x 9
Overall Strategy
- Resample T2 reference to isotropic voxel size
- Manually reorient DWI into orientation of T2
- Resample DWI into new space and into isotropic voxel size
- Convert DWI -> DTI, incl. mask & baseline extraction
- Register DTI_baseline to T2 (affine+nonrigid) w/o masking
- Resample DTI_mask with above transform to build mask for T2
- Repeat registration above, this time incl. masks and with higher BSpline grid size
- Resample DTI with above transform
Procedures Part I : Preprocessing
- Slicer 4.1 recommended modules: BrainsFit, Resample DTI Volume, Diffusion Tensor Estimation
- Note: if not interested in the preprocessing steps, download only the main dataset and skip phase I-III.
- Phase I: resample T2 to isotropic voxel size
- original T2 has 0.5 x 0.5 x 3 mm voxel size, which we resample to 1.2 x 1.2 x 1.2 mm (you may set 1x1x1 if you have sufficient RAM)
- open Modules:AllModules:ResampleScalarVolume module
- Spacing: enter 1,1,1
- Interpolation: check hamming
- Input Volume: "T2_raw"
- Output Volume: create new volume, rename to "T2_iso"
- click: Apply
- Save generated volume as "T2_iso.nrrd"
- Phase IIa: Build DWI mask + baseline
- open the Modules:Diffusion:DiffusionWeightedImages:DiffusionWeightedVolumeMasking module
- Input DWI Volume: "DWI_raw"
- Output Baseline Volume: Create New Volume, rename to "DWI_baseline"
- Output Threshold Mask: Create New Volume, rename to "DWI_mask"
- Leave other settings at default; click Apply
- Save generated volume as "DWI_mask.nrrd"
- Phase IIb: Build Masks: T2 mask
- open the Modules:Segmentation:SimpleRegionGrowing module
- set Smoothing iterations : 0
- set number of iterations : 1
- set Multiplier: 0.8
- set Neighborhood Radius : 5
- set Output Label Value : 1
- set Seeds: "create New fiducial List"
- in toolbar select Fiducial mouse mode. Then place 2 fiducials in the center of the brain within the deep gray or deep WM (picture). Avoid selecting CSF.
- Input Volume": "T2_iso"
- Output Volume: "T2_iso_mask"
- Click Apply. Upon completion you should get a sparse segmentation of the WM (see picture)
- go to Modules:Editor module
- if not chosen by default, select "T2_iso_mask" as labelmap to edit
- Select Dilation tool. Click Apply button several times (4-5x) until most of the brain is filled
- Select brush tool, adjust radius, set label to 0 and remove portions extending beyond the brain. Do not worry about holes in CSF not filled or small regions, they will not affect the registration
- Open the Modules:Filtering:Voting Binary Hole Filling Image Filter module
- Maximum Radius: 7,7,7
- Majority Threshold: 1
- Background: 0 ; Foreground: 1
- Input Volume: T2_iso_mask
- Output Volume: T2_iso_mask
- click Apply.
- Save generated map as "T2_iso_mask.nrrd"
- Phase III: Register DWI to T2_iso
- open the Registration : Modules:Registration:General Registration (BRAINS) module
- Input Images: fixed = T2_iso , moving = DWI_baseline
- Output Settings:
- Slicer BSpline Transform (create new transform, rename to: "Xf1_DWbase-T2_BSpline")
- Slicer Linear Transform none
- Output Image Volume (create new volume, rename to: "DWI_baseline_Xf1"
- Registration Phases: select/check Rigid , Rigid+Scale, Affine, BSpline
- Main Parameters:
- increase Number Of Samples to 200,000
- set B-Spline Grid Size to 5,5,5
- Mask Option:
- select ROI radio button
- ROI input fixed: "T2_iso_mask" (from phase IIb above)
- ROI input moving: "DWI_mask" (from phase IIa above)
- Leave all other settings at default
- click: Apply; runtime < 1-3 min.
- Phase III: Resample DTI'
- Open the Resample DTI Volume module (found under: All Modules)
- Input Volume: select DTI
- Output Volume: select create new Diffusion Tensor Volume,and rename it to DTI_Xf1
- Reference Volume: select T2
- Transform Parameters: select transform node "Xf1_DTI-T2_BSpline", for Deformation Field: none ; check the displacement checkbox
- Leave all other settings at defaults
- Click Apply; runtime ~ 2 min.
- set T2 as background and new DTI_Xf1 volume as foreground
- fade between back- and foreground to see DTI overlay onto the T2 image. Note that you can also fade via holding the OPTION+CMD keys (mac) + dragging left mouse.
- Phase III: DWI -> DTI'
- open "Diffusion Tensor Estimation" module
- Input DWI Volume: DWI_Xf0, Output DTI Volume: create new, rename to "DTI"
- Output Baseline: create new, rename to "DTI_base"
- Otsu Threshold Mask: create new, rename to "DTI_mask"
- Estimation Parameters: reduce Otsu Omega Threshold" to 0.3
- check boxes for Remove Islands and Apply Mask
- Click: Apply
Procedures Part II : Registration
- Phase IV: Register DTI (unmasked)
- open Registration : General Registration (BRAINS) module
- Input Images: fixed = T2 , moving = DTI_base
- Output Settings:
- Slicer BSpline Transform (create new transform, rename to: "Xf1_DTbase-T2_BSpline")
- Slicer Linear Transform none
- Output Image Volume (create new volume, rename to: "DTIbaseline_Xf1"
- Registration Phases: select/check Rigid , Rigid+Scale, Affine, BSpline
- Main Parameters:
- increase Number Of Samples to 200,000
- set B-Spline Grid Size to 5,5,5
- Leave all other settings at default
- click: Apply; runtime < 1 min.
- Phase V: Resample DTI_mask (presets: DTI_mask_Xf1)
- we use the above Xform to produce a mask for the T2.
- Open Filtering / Resample Scalar/Vector/DWI module
- Input Volume: DTI_mask; Output volume: create new volume, rename to "DTI_mask_Xf1"
- Transform Node: "Xf1_DTI-T2_unmasked"
- select/check: output-to-input
- Interpolation Type: select: nn
- click: Apply
- Go to Volumes module, select the new "DTI_mask_Xf1", in the Info tab, check the Labelmap box
- Phase VI:REGISTER DTI TO T2 with masking
- open Registration : BrainsFit module (presets: Xf2_DTI-T2_masked)
- set T2_Xf1 as fixed and DTI_baseline as moving image
- select/check Include Rigid registration phase
- select/check Include ScaleVersor3D registration phase
- select/check Include Affine registration phase
- select/check Include BSpline registration phase
- Output BSpline Transform: create new , rename to "Xf2_DTI-T2_masked"
- Output Volume: create new, rename to "DTI_base_Xf2"
- Registration Parameters: increase Number Of Samples to 200,000
- Registration Parameters: set Number Of Grid Subdivisions to 7,7,5
- Control of Mask Processing
- select/check: ROI (rightmost box)
- Input Fixed Mask: select "DTI_mask_Xf1"
- Input Moving Mask: select "DTI_mask"
- Leave all other settings at default
- click: Apply; runtime < 1 min.
- set T2_Xf1 as fixed and DTI_baseline as moving image
- Phase VII: Resample DTI
- Load the combined transform (Add Data)
- Open the Resample DTI Volume module (found under: All Modules)
- Input Volume: select DTI
- Output Volume: select New DTI Volume, rename to DTI_Xf2
- Reference Volume: select T2
- Transform Parameters: select transform "Xf2_DTI-T2_masked
- check box: output-to-input
- Leave all other settings at defaults
- Click Apply; runtime ~ 2 min.
- Go to the Volumes module, select the newly produced DTI_Xf2 volume
- under the Display tab, select Color Orientation from the Scalar Mode menu
- set T1 as background and new DTI_Xf2 volume as foreground
- Set fade slider to see DTI overlay onto the T2 image
for more details see the tutorial(s) under Downloads above
Registration Results
| baseline to T2 after affine+nonrigid alignment |
Discussion: Key Strategies
- the two images have identical contrast, hence we could consider "sharper" cost functions, such as NormCorr or MeanSqrd. But because of the strong distortions and lower resolution of the moving image, Mutual Information is recommended as the most robust metric.
- often anatomical labels are available from the reference scan. It would be less work to align the anatomical reference with the DTI, since that would circumvent having to resample the complex tensor data into a new orientation. However the strong distortions are better addressed by registering the other direction, i.e. move the DTI into the anatomical reference space.
- in this example the initial alignment of the two scans is very poor. The strongly oblique orientation of the DWI makes an initial manual alignment step necessary. This step should occur before converting to the DTI to avoid interpolation artifacts.