Difference between revisions of "Algorithm:BU"
| (87 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
Back to [[Algorithm:Main|NA-MIC Algorithms]] | Back to [[Algorithm:Main|NA-MIC Algorithms]] | ||
__NOTOC__ | __NOTOC__ | ||
| − | = Overview of | + | = Overview of SUNY Stony Brook Algorithms (PI: Allen Tannenbaum) = |
| − | At | + | At the State University of New York at Stony Brook, we are broadly interested in a range of mathematical image analysis algorithms for segmentation, registration, diffusion-weighted MRI analysis, and statistical analysis. For many applications, we cast the problem in an energy minimization framework wherein we define a partial differential equation whose numeric solution corresponds to the desired algorithmic outcome. The following are many examples of PDE techniques applied to medical image analysis. |
| − | = | + | = Stony Brook University Projects = |
{| cellpadding="10" style="text-align:left;" | {| cellpadding="10" style="text-align:left;" | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:LiverFibrosisHist.png|200px|]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:LiverFibrosisStaging|Liver Fibrosis Staging by MRI Analysis]] == | ||
| + | |||
| + | In this project, we provide tools for robust liver fibrosis staging, based on MRI image analysis. The current | ||
| + | practice of fibrosis assessment, which is based on painful liver biopsy, might be dangerous. | ||
| + | Moreover, the decision of the pathologist based on a biopsy is subjective, and depends | ||
| + | on the sample, because the fibrosis level varies along the liver. No objective standard has | ||
| + | been developed yet for histological fibrosis assessment. Magnetic resonance volume data | ||
| + | has much lower resolution than histological image data, but it includes the entire liver | ||
| + | volume. Also, MRI is non-invasive and not painful, thus it is preferred as a diagnostic tool. | ||
| + | Previously it has been hypothesized that the average brightness of Apparent Diffusion Coefficient (ADC) | ||
| + | in diffusion MRI correlates with the fibrosis stage. [[Projects:LiverFibrosisStaging|More...]] | ||
| + | |||
| + | |- | ||
| + | |||
| + | |||
| + | | | [[Image:Heart_topology.jpg|200px|]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:TopologicalSegmentation|Left Atrium Wall Segmentation Using Topological Features]] == | ||
| + | |||
| + | Catheter ablation has been proposed for treatment of atrial fibrillation arrhythmia. MRI | ||
| + | data, obtained at University of Utah, are used to explore lesion ablation and scarification | ||
| + | locations. In addition, MRI analysis may help to predict if the ablation procedure will help | ||
| + | a patient or not. Many of these image analysis tasks are largely based on segmentation of | ||
| + | left atrial wall, which is done manually or semi-automatically. Automatic segmentation uses | ||
| + | moving contours or surfaces (interfaces) to segment image data by minimizing a predefined | ||
| + | energy function. These moving interfaces are highly affected by image data, which can be | ||
| + | thought as a force field pushing the interface to features of choice. Thus, the choice of | ||
| + | interface attracting image features is critical. [[Projects:TopologicalSegmentation|More...]] | ||
| + | |||
| + | |- | ||
| + | |||
| | [[Image:toT1e1.png|200px|]] | | | [[Image:toT1e1.png|200px|]] | ||
| | | | | | ||
| − | == [[Projects:RegistrationTBI|Multimodal Deformable Registration of Traumatic Brain Injury MR Volumes | + | == [[Projects:RegistrationTBI|Multimodal Deformable Registration of Traumatic Brain Injury MR Volumes]] == |
| − | Time-efficient processing and analysis of magnetic resonance imaging (MRI) volumes | + | Time-efficient processing and analysis of magnetic resonance imaging (MRI) volumes is desirable for the neurocritical care and monitoring of traumatic brain injury (TBI) patients. An important problem of TBI neuroimaging data analysis is the task of co-registering MR volumes acquired using distinct sequences in the presence of widely variable pixel intensities that are due to the presence of pathology. Here we address this important and challenging problem using an implementation of multimodal deformable registration methods. One method have been implemented on graphics processing units (GPU). In this method we follow a viscous fluid model framework and replace mutual information with the Bhattacharyya distance as the measure of similarity between image volumes. The proposed algorithm is implemented on a GPU and its robustness is illustrated using a longitudinal multimodal TBI dataset. Another method proposes an extension |
| + | to the principal axis transformation method for finding robust rigid transformation of two | ||
| + | volumes. The additional elastic registration is based on a volume registration method | ||
| + | MIND, proposed recently by Heinrich et al. [[Projects:RegistrationTBI|More...]] | ||
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:HandTracking.png|200px|]] |
| | | | | | ||
| − | == [[Projects: | + | == [[Projects:SobolevTracker|Object Tracking With Adaptive Sobolev Active Contours]] == |
| − | + | In this project we propose adaptive tracking mechanism which can be used in medical video applications, or 3D volume segmentation. The proposed Sobolev active contour model overcomes the | |
| + | problems of occlusions and changes in scale by adaptive tweaking of the rigidity parameters. The proposed tracking algorithms work in a variety of scenarios and deal naturally with | ||
| + | clutter and noise in the scenes, object deformations, partial and entire object occlusions, and | ||
| + | low contrast objects. Experimental results show the advantages of our approach compared | ||
| + | to state-of-the-art visual trackers.[[Projects:SobolevTracker|More...]] | ||
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:MultiScaleHippoSegmentationHausdorf.png|200px]] |
| | | | | | ||
| − | == [[Projects: | + | == [[Projects:MultiScaleShapeSegmentation|Multi-scale Shape Representation, Registration, and Segmentation With Applications to Radiotherapy]] == |
| + | |||
| + | We present in this work a multiscale representation for shapes with arbitrary topology, and a method to segment the target organ/tissue from medical images having very low contrast with respect to surrounding regions using multiscale shape information and local image features. In a number of previous papers, shape knowledge was incorporated by first constructing a shape space from training data, and then constraining the segmentation process to be within the resulting shape space. However, such an approach has certain limitations including the fact that small scale shape variances may be overwhelmed by those on larger scale, and therefore the local shape information is lost. In this work, first we handle this problem by providing a multiscale shape representation using the wavelet transform. Consequently, the shape variances captured by the statistical learning step are also represented at various scales. In doing so, not only is the diversity of shape enriched, but also small scale changes are nicely captured. [[Projects:MultiScaleShapeSegmentation|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> Gao, Y. and Corn, B. and Schifter, D. and Tannenbaum, A. Multiscale 3D Shape Representation and Segmentation with Applications to Hippocampal/Caudate Extraction from Brain MRI, Medical Image Analysis. 16(2) pp374, 2012 | ||
| − | |||
| − | |||
|- | |- | ||
| Line 43: | Line 90: | ||
| | | | | | ||
| − | == [[Projects: | + | == [[Projects:AFibSegmentationRegistration|Segmentation and Registration for Atrial Fibrillation Ablation Therapy]] == |
Magnetic resonance imaging (MRI) has been used for both pre- and and post-ablation assessment of the atrial wall. MRI can aid in selecting the right candidate for the ablation procedure and assessing post-ablation scar formations. Image processing techniques can be used for automatic segmentation of the atrial wall, which facilitates an accurate statistical assessment of the region. As a first step towards the general solution to the computer-assisted segmentation of the left atrial wall, in this research we propose a shape-based image segmentation framework to segment the endocardial wall of the left atrium.[[Projects:SegmentationEpicardialWall|More...]] | Magnetic resonance imaging (MRI) has been used for both pre- and and post-ablation assessment of the atrial wall. MRI can aid in selecting the right candidate for the ablation procedure and assessing post-ablation scar formations. Image processing techniques can be used for automatic segmentation of the atrial wall, which facilitates an accurate statistical assessment of the region. As a first step towards the general solution to the computer-assisted segmentation of the left atrial wall, in this research we propose a shape-based image segmentation framework to segment the endocardial wall of the left atrium.[[Projects:SegmentationEpicardialWall|More...]] | ||
| Line 72: | Line 119: | ||
expert and non-expert human examiners.[[Projects:PainAssessment|More...]] | expert and non-expert human examiners.[[Projects:PainAssessment|More...]] | ||
| − | <font color="red">'''New: '''</font> B. Gholami, W. M. Haddad, and A. Tannenbaum, | + | <font color="red">'''New: '''</font> B. Gholami, W. M. Haddad, and A. Tannenbaum, Relevance Vector Machine Learning for Neonate Pain Intensity Assessment Using Digital Imaging. IEEE Trans. Biomed. Eng., vol. 57, pp. 1457-1466, 2012. |
| − | B. Gholami, W. M. Haddad, and A. R. Tannenbaum, | + | <font color="red">'''New: '''</font> B. Gholami, W. M. Haddad, and A. R. Tannenbaum, Agitation and Pain Assessment Using Digital Imaging. Proc. IEEE Eng. Med. Biolog. Conf., Minneapolis, MN, pp. 2176-2179, 2009 (Awarded National Institute of Biomedical Imaging and Bioengineering/National Institute of Health Student Travel Fellowship). |
| + | <font color="red">'''New: '''</font> Wassim M. Haddad, James M. Bailey, Behnood Gholami, and Allen Tannenbaum. Optimal Drug Dosing Control for Intensive Care Unit Sedation Using a Hybrid Deterministic-Stochastic Pharmacokinetic and Pharmacodynamic | ||
| + | Model. Optimal Control, Applications and Methods}, 2012, DOI: 10.1002/oca.2038. | ||
|- | |- | ||
| Line 86: | Line 135: | ||
Multiple objects are segmented simultaneously using several interactive active contours based on the feature image which utilizes the robust statistics of the image. [[RobustStatisticsSegmentation|More...]] | Multiple objects are segmented simultaneously using several interactive active contours based on the feature image which utilizes the robust statistics of the image. [[RobustStatisticsSegmentation|More...]] | ||
| − | + | <font color="red">'''New: '''</font> Y. Gao, S. Bouix, M. Shenton, R. Kikinis, A. Tannenbaum. A 3D interactive multi-object segmentation tool using local robust statistics driven active contours. MedIA, volume 16, 2012, pp. 1216-1227. | |
|- | |- | ||
| Line 96: | Line 145: | ||
The 3D prostate MRI images are collected by collaborators at Queen’s University. With a little manual initialization, the algorithm provided the results give to the left. The method mainly uses Random Walk algorithm. [[Projects:ProstateSegmentation|More...]] | The 3D prostate MRI images are collected by collaborators at Queen’s University. With a little manual initialization, the algorithm provided the results give to the left. The method mainly uses Random Walk algorithm. [[Projects:ProstateSegmentation|More...]] | ||
| − | <font color="red">'''New: '''</font> Y. Gao, A. Tannenbaum | + | <font color="red">'''New: '''</font> Y. Gao, R. Sandhu, G. Fichtinger, A. Tannenbaum. A coupled global registration and segmentation framework with application to magnetic resonance prostate imagery. IEEE Trans. Medical Imaging, volume 29, 2010, pp. 1781-1794. |
| − | |||
|- | |- | ||
| Line 108: | Line 156: | ||
3D volumetric image registration is performed. The method is based on registering the images through point sets, which is able to handle long distance between as well as registration between Supine and Prone pose prostate. [[Projects:pfPtSetImgReg|More...]] | 3D volumetric image registration is performed. The method is based on registering the images through point sets, which is able to handle long distance between as well as registration between Supine and Prone pose prostate. [[Projects:pfPtSetImgReg|More...]] | ||
| − | <font color="red">'''New: '''</font> Y. Gao, R. Sandhu, G. Fichtinger, A. Tannenbaum; A coupled global registration and segmentation framework with application to magnetic resonance prostate imagery. IEEE TMI vol.29, | + | <font color="red">'''New: '''</font> Y. Gao, R. Sandhu, G. Fichtinger, A. Tannenbaum; A coupled global registration and segmentation framework with application to magnetic resonance prostate imagery. IEEE TMI vol.29, 2010, pp. 1781-1794. |
| − | . | + | |
| + | <font color="red">'''New: '''</font> Y. Gao, Y. Rathi, S. Bouix, A. Tannenbaum; Filtering in the diffeomorphism group and the registration of point sets. IEEE Transactions Image Processing, vol. 21, 2012, pp. 4383-4396 . | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[File:LASegAxialView.png|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:LeftAtriumSegmentation|Left Atrium Segmentation for Atrial Fibrillation Treatment]] == | ||
| + | |||
| + | The planning and evaluation of left atrial ablation procedures is commonly based on the segmentation of the left atrium, which is a challenging task due to large anatomical | ||
| + | variations. In this paper, we propose an automatic approach for segmenting the left atrium from magnetic resonance imagery (MRI). The segmentation problem is formulated as a problem in variational region growing. [[Projects:LeftAtriumSegmentation|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> L. Zhu, Y. Gao, A. Yezzi, A. Tannenbaum. Automatic Segmentation of the Left Atrium from MRI images using Variational Region Growing with a Shape Prior, IEEE Transaction on Image Processing, vol. 22, no. 12, 2013. | ||
| + | |||
| + | <font color="red">'''New: '''</font>L. Zhu, Y. Gao, A. Yezzi, R. MacLeod, J. Cates, A. Tannenbaum. Automatic Segmentation of the Left Atrium from MRI Images Using Salient Feature and Contour Evolution, IEEE Engineering in Medicine and Biology Conference(EMBC), 2012. | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:ScarSeg_EM.png|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:ScarIdentification|Scar Tissue Identification for Post-Ablation Analysis]] == | ||
| + | The delay-enhanced MRI (DE-MRI) technique provides an effective way of imaging scarring and fibrosis tissue of atria. Segmentation of the LA from DE-MRI images can | ||
| + | be used in atrial fibrillation (AF) treatment to select suitable candidates for ablation therapy and subsequent monitoring of the therapy. [[Projects:ScarIdentification|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> Y. Gao, L. Zhu, A. Yezzi, S. Bouix , A. Tannenbaum. Scar Segmentation in DE-MRI, IEEE International Symposium on Biomedical Imaging (ISBI) , 2012. | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:LongitudinalAFib.png|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:AFibLongitudinalAnalysis|Longitudinal Shape Analysis for AFib]] == | ||
| + | The shape evolution of the left atrium in the atrial fibrillation patiens is studied longitudinally to reveal the difference between recover group and the AFib recurrence group. [[Projects:AFibLongitudinalAnalysis|More...]] | ||
|- | |- | ||
| − | | | [[Image:GT-DWI-Reorientation-1.jpg| | + | | | [[Image:LRV_Wall.png|200px]] |
| + | | | | ||
| + | |||
| + | == [[Projects:VentricleSegmentation|Ventricles Segmentation for Diagnosis of Cardiac Diseases]] == | ||
| + | This work presents an automatic method for extracting the myocardial wall of the left and right ventricles from cardiac CT images. In the method, the left and right ven- | ||
| + | tricles are located sequentially, in which each ventricle is detected by first identifying the endocardial surface and then segmenting the epicardial surface. [[Projects:VentricleSegmentation|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> L. Zhu, Y. Gao, V. Appia, A. Yezzi, C. Arepalli , A. Stillman, and A. Tannenbaum. Automatic Extraction of the Myocardial Wall from CT Images using Shape Segmentation and Variational Region Growing, IEEE Transaction on Biomedical Engineering, vol. 60, no. 10, 2013. | ||
| + | |- | ||
| + | |||
| + | | | [[Image:RiskMassSeg.png|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:RiskMassEstimation|Risk Mass Estimation for Heart Risk Evaluation]] == | ||
| + | Prognosis and treatment of cardiovascular diseases frequently require the determination of the myocardial mass at risk caused by coronary stenoses. However, few work has been done for estimating the myocardial mass at risk directly from the heart surface segmented from CAT imagery, rather than using a simplified heart model such as ellipsoid. [[Projects:RiskMassEstimation|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> L. Zhu, Y. Gao, A. Yezzi, C. Arepalli , A. Stillman, and A. Tannenbaum. A Computational Framework for Estimating the Mass at Risk Caused by Stenoses using CT Angiography, Internatial Journal of Cardiac Imaging(IJCI), In preparation. | ||
| + | |||
| + | <font color="red">'''New: '''</font> L. Zhu, Y. Gao, V. Mohan, A. Stillman, T. Faber, A. Tannenbaum. Estimation of myocardial volume at risk from CT angiography, Proceedings of SPIE , pp.79632-38A, 2011. | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:GT-DWI-Reorientation-1.jpg|300px]] | ||
| | | | | | ||
| Line 120: | Line 224: | ||
This work proposes a methodology to segment tubular fiber bundles from diffusion weighted magnetic resonance images (DW-MRI). Segmentation is simplified by locally reorienting diffusion information based on large-scale fiber bundle geometry. [[Projects:DWIReorientation|More...]] | This work proposes a methodology to segment tubular fiber bundles from diffusion weighted magnetic resonance images (DW-MRI). Segmentation is simplified by locally reorienting diffusion information based on large-scale fiber bundle geometry. [[Projects:DWIReorientation|More...]] | ||
| − | <font color="red">'''New: '''</font> Near- | + | <font color="red">'''New: '''</font> Near-Tubular Fiber Bundle Segmentation for Diffusion Weighted Imaging: Segmentation Through Frame Reorientation. Neuroimage, volume 45, 2009, pp. 123-132. |
|- | |- | ||
| Line 131: | Line 235: | ||
We have proposed a new model for tubular surfaces that transforms the problem of detecting a surface in 3D space, to detecting a curve in 4D space. Besides allowing us to impose a "soft" tubular shape prior, this also leads to computational efficiency over conventional surface segmentation approaches. [[Projects:TubularSurfaceSegmentation|More...]] | We have proposed a new model for tubular surfaces that transforms the problem of detecting a surface in 3D space, to detecting a curve in 4D space. Besides allowing us to impose a "soft" tubular shape prior, this also leads to computational efficiency over conventional surface segmentation approaches. [[Projects:TubularSurfaceSegmentation|More...]] | ||
| + | <font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi, A. Stillman and A. Tannenbaum. Vessel Segmentation with Automatic Centerline Extraction using Tubular Surface Segmentation. September 2009. Proceedings of the Workshop on Cardiac Interventional Imaging and Biophysical Modelling (CI2BM'09), Int Conf Med Image Comput Comput Assist Interv. 2009. | ||
| − | <font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi | + | <font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi and A. Tannenbaum. Tubular Surface Segmentation for Extracting Anatomical Structures from Medical Imagery, IEEE Transactions on Medical Imaging, volume 29, 2011, pp. 1945-1958. |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| Line 150: | Line 252: | ||
<font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi, M. Kubicki, D. Terry and A. Tannenbaum. Population Analysis of the Cingulum Bundle using the Tubular Surface Model for Schizophrenia Detection. SPIE Medical Imaging 2010. | <font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi, M. Kubicki, D. Terry and A. Tannenbaum. Population Analysis of the Cingulum Bundle using the Tubular Surface Model for Schizophrenia Detection. SPIE Medical Imaging 2010. | ||
| − | <font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi, M. Kubicki and A. Tannenbaum. Population Analysis of neural fiber bundles towards schizophrenia detection and characterization, using the Tubular Surface model. Neuroimage (in | + | <font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi, M. Kubicki and A. Tannenbaum. Population Analysis of neural fiber bundles towards schizophrenia detection and characterization, using the Tubular Surface model. Neuroimage (in submission) |
| + | |||
| + | |||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:KVoutSegTightMod.png|200px]] | ||
| + | | | | ||
| + | |||
| + | |||
| + | == [[Projects:InteractiveSegmentation|Interactive Image Segmentation With Active Contours]] == | ||
| + | |||
| + | An approach for tightly coupling the user into a semi-automatic segmentation framework is proposed in this work. A human guides the automatic segmentation by iteratively providing input until convergence to the desired segmentation. The result is a segmentation of manual quality in a fraction of the time; the whole process is intuitive and highly flexible [[Projects:InteractiveSegmentation|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> I. Kolesov, P.Karasev, G.Muller, K.Chudy, J.Xerogeanes, and A. Tannenbaum. Human Supervisory Control Framework for Interactive Medical Image Segmentation. MICCAI Workshop on Computational Biomechanics for Medicine 2011. | ||
| + | |||
| + | |||
| + | <font color="red">'''New: '''</font> P.Karasev, I.Kolesov, K.Chudy, G.Muller, J.Xerogeanes, and A. Tannenbaum. Interactive MRI Segmentation with Controlled Active Vision. IEEE CDC-ECC 2011. | ||
| + | |||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:PostRegFleshSkeleton.png|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:MGH-HeadAndNeck-PtSetReg|Constrained Registration for Adaptive Radiotherapy]] == | ||
| + | |||
| + | A hierarchical approach is described to register two CT scans from different patients. The registration process extracts point clouds representing anatomical structures and aligns them sequentially. The proposed method for registering point clouds can incorporate a variety of constraints including restriction on the injectivity of the deformation field and stationarity of selected landmarks. [[Projects:MGH-HeadAndNeck-PtSetReg|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> I. Kolesov, J. Lee, P.Vela, G. Sharp and A. Tannenbaum. Diffeomorphic Point Set Registration with Landmark Constraints. In Preparation for PAMI. | ||
| + | |||
| Line 159: | Line 291: | ||
| | | | | | ||
| − | == [[Projects:MGH-HeadAndNeck-RT|Adaptive Radiotherapy for | + | == [[Projects:MGH-HeadAndNeck-RT|Adaptive Radiotherapy for Head, Neck and Thorax]] == |
We proposed an algorithm to include prior knowledge in previously segmented anatomical structures to help in the segmentation of the next structure. This will add enough prior information to allow the Graph Cuts algorithm to segment structures with fuzzy boundaries. [[Projects:MGH-HeadAndNeck-RT|More...]] | We proposed an algorithm to include prior knowledge in previously segmented anatomical structures to help in the segmentation of the next structure. This will add enough prior information to allow the Graph Cuts algorithm to segment structures with fuzzy boundaries. [[Projects:MGH-HeadAndNeck-RT|More...]] | ||
| Line 172: | Line 304: | ||
| | | | | | ||
| − | == [[Projects:KPCASegmentation|Kernel | + | == [[Projects:KPCASegmentation|Kernel Methods for Segmentation]] == |
| − | Segmentation performances using active contours can be drastically improved if the possible shapes of the object of interest are learnt. The goal of this work is to use Kernel PCA to learn shape priors. Kernel PCA allows for learning | + | Segmentation performances using active contours can be drastically improved if the possible shapes of the object of interest are learnt. The goal of this work is to use Kernel PCA to learn shape priors. Kernel PCA allows for learning nonlinear dependencies in data sets, leading to more robust shape priors. [[Projects:KPCASegmentation|More...]] |
| − | <font color="red">'''New: '''</font> S. Dambreville | + | <font color="red">'''New: '''</font> R. Sandhu, S. Dambreville, and A. Tannenbaum. A Non-Rigid Kernel Based Framework for 2D/3D Pose Estimation and 2D Image Segmentation. IEEE Trans Pattern Anal Mach Intelligence, volume 33, 2011, pp. 1098-1115. |
|- | |- | ||
| Line 186: | Line 318: | ||
In this work, we provide an energy minimization framework which allows one to find fiber tracts and volumetric fiber bundles in brain diffusion-weighted MRI (DW-MRI). [[Projects:GeodesicTractographySegmentation|More...]] | In this work, we provide an energy minimization framework which allows one to find fiber tracts and volumetric fiber bundles in brain diffusion-weighted MRI (DW-MRI). [[Projects:GeodesicTractographySegmentation|More...]] | ||
| − | <font color="red">'''New: '''</font> J. Melonakos, E. Pichon, S. | + | <font color="red">'''New: '''</font> J. Melonakos, E. Pichon, S. Angenent, and A. Tannenbaum. Finsler Active Contours. IEEE Transactions on Pattern Analysis and Machine Intelligence, volume 30, 2008, pp. 412-423. |
|- | |- | ||
| Line 197: | Line 329: | ||
Many techniques for multi-shape representation may often develop inaccuracies stemming from either approximations or inherent variation. Label space is an implicit representation that offers unbiased algebraic manipulation and natural expression of label uncertainty. We demonstrate smoothing and registration on multi-label brain MRI. [[Projects:LabelSpace|More...]] | Many techniques for multi-shape representation may often develop inaccuracies stemming from either approximations or inherent variation. Label space is an implicit representation that offers unbiased algebraic manipulation and natural expression of label uncertainty. We demonstrate smoothing and registration on multi-label brain MRI. [[Projects:LabelSpace|More...]] | ||
| − | <font color="red">'''New: '''</font> J. Malcolm, Y. Rathi, A. Tannenbaum. | + | <font color="red">'''New: '''</font> J. Malcolm, Y. Rathi, S. Bouix, M. Shenton, A. Tannenbaum. Affine registration of label maps in label space. Journal of Computing, volume 2, 2010, pp, 1-11. |
|- | |- | ||
| Line 208: | Line 340: | ||
High accuracy imaging and image processing techniques allow for collecting structural information of biomolecules with atomistic accuracy. Direct interpretation of the dynamics and the functionality of these structures with physical models, is yet to be developed. Clustering of molecular conformations into classes seems to be the first stage in recovering the formation and the functionality of these molecules. [[Projects:NonParametricClustering|More...]] | High accuracy imaging and image processing techniques allow for collecting structural information of biomolecules with atomistic accuracy. Direct interpretation of the dynamics and the functionality of these structures with physical models, is yet to be developed. Clustering of molecular conformations into classes seems to be the first stage in recovering the formation and the functionality of these molecules. [[Projects:NonParametricClustering|More...]] | ||
| − | <font color="red">'''New: '''</font> E. Hershkovits, A. Tannenbaum, | + | <font color="red">'''New: '''</font> X. LeFaucheur, E. Hershkovits, R. Tannenbaum, and A. Tannenbaum. Non-parametric clustering for studying RNA conformations. IEEE Trans. Computational Biology and Bioinformatics, volume 8, 2011, pp. 1604-1618. |
| + | |||
| + | <font color="red">'''New: '''</font> Il Tae Kim, A. Tannenbaum, R. Tannenbaum. Anisotropic conductivity of magnetic carbon nanotubes | ||
| + | embedded in epoxy matrices. Carbon, volume 49, 2011, pp. 54-61. | ||
|- | |- | ||
| Line 220: | Line 355: | ||
a rigid body transformation. Experimental results are provided that demonstrate the robustness of the algorithm to initialization, noise, missing structures or differing point densities in each sets, on challenging 2D and 3D registration tasks. [[Projects:PointSetRigidRegistration|More...]] | a rigid body transformation. Experimental results are provided that demonstrate the robustness of the algorithm to initialization, noise, missing structures or differing point densities in each sets, on challenging 2D and 3D registration tasks. [[Projects:PointSetRigidRegistration|More...]] | ||
| − | <font color="red">'''New: '''</font> R. Sandhu, S. Dambreville, A. Tannenbaum. | + | <font color="red">'''New: '''</font> R. Sandhu, S. Dambreville, A. Tannenbaum. Point set registration via particle filtering and stochastic dynamics. IEEE TPAMI, volume 32, 2010, pp. 1459-1473. |
| + | <font color="red">'''New: '''</font> R. Sandhu, S. Dambreville, A. Tannenbaum. A non-rigid kernel based framework for 2D/3D pose estimation and 2D image segmentation. IEEE TPAMI, volume 33, 2011, pp. 1098-1115. | ||
|- | |- | ||
| Line 228: | Line 364: | ||
| | | | | | ||
| − | == [[Projects:OptimalMassTransportRegistration|Optimal Mass Transport Registration]] == | + | == [[Projects:OptimalMassTransportRegistration|Optimal Mass Transport Registration and Visualization]] == |
| + | |||
| + | The goal of this project is to implement a computationally efficient Elastic/Non-rigid Registration algorithm based on the Monge-Kantorovich theory of optimal mass transport for 3D Medical Imagery. Our technique is based on Multigrid and Multiresolution techniques. This method is particularly useful because it is parameter free and utilizes all of the grayscale data in the image pairs in a symmetric fashion and no landmarks need to be specified for correspondence. [[Projects:OptimalMassTransportRegistration|More...]] | ||
| − | + | <font color="red">'''New: '''</font> Eldad Haber, Tauseef Rehman, and Allen Tannenbaum. An Efficient Numerical Method for the Solution of the L2 Optimal Mass Transfer Problem. SIAM Journal of Scientific Computing, volume 32, 2011, pp. 197-211. | |
| − | <font color="red">'''New: '''</font> Eldad Haber, | + | <font color="red">'''New: '''</font> Tauseef Rehman, Eldad Haber, Gallagher Pryor, and Allen Tannenbaum. 3D nonrigid registration via optimal mass transport on the GPU. MedIA, volume 13, 2010, pp. 931-940. |
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> Ayelet Dominitz and Allen Tannenbaum. Texture Mapping Via Optimal Mass Transport. IEEE TVCG, volume 16, 2010), pp. 419-433. |
|- | |- | ||
| Line 244: | Line 382: | ||
To represent multiscale variations in a shape population in order to drive the segmentation of deep brain structures, such as the caudate nucleus or the hippocampus. [[Projects:MultiscaleShapeSegmentation|More...]] | To represent multiscale variations in a shape population in order to drive the segmentation of deep brain structures, such as the caudate nucleus or the hippocampus. [[Projects:MultiscaleShapeSegmentation|More...]] | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:GT-SPD-img1.png|200px|]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:SoftPlaqueDetection|Soft Plaque Detection in CTA Imagery]] == | ||
| + | |||
| + | The ability to detect and measure non-calcified plaques (also known as soft plaques) may improve physicians’ ability to predict cardiac events. This work automatically detects soft plaques in CTA imagery using active contours driven by spatially localized probabilistic models. Plaques are identified by simultaneously segmenting the vessel from the inside-out and the outside-in using carefully chosen localized energies [[Projects:SoftPlaqueDetection|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> Soft Plaque Detection and Automatic Vessel Segmentation. PMMIA Workshop in MICCAI, Sep. 2009. | ||
|- | |- | ||
| Line 286: | Line 435: | ||
| | | | | | ||
| − | == [[Projects:ConformalFlatteningRegistration|Conformal Flattening | + | == [[Projects:ConformalFlatteningRegistration|Conformal Surface Flattening]] == |
The goal of this project is for better visualizing and computation of neural activity from fMRI brain imagery. Also, with this technique, shapes can be mapped to shperes for shape analysis, registration or other purposes. Our technique is based on conformal mappings which map genus-zero surface: in fmri case cortical or other surfaces, onto a sphere in an angle preserving manner. [[Projects:ConformalFlatteningRegistration|More...]] | The goal of this project is for better visualizing and computation of neural activity from fMRI brain imagery. Also, with this technique, shapes can be mapped to shperes for shape analysis, registration or other purposes. Our technique is based on conformal mappings which map genus-zero surface: in fmri case cortical or other surfaces, onto a sphere in an angle preserving manner. [[Projects:ConformalFlatteningRegistration|More...]] | ||
Latest revision as of 01:56, 5 June 2014
Home < Algorithm:BUBack to NA-MIC Algorithms
Overview of SUNY Stony Brook Algorithms (PI: Allen Tannenbaum)
At the State University of New York at Stony Brook, we are broadly interested in a range of mathematical image analysis algorithms for segmentation, registration, diffusion-weighted MRI analysis, and statistical analysis. For many applications, we cast the problem in an energy minimization framework wherein we define a partial differential equation whose numeric solution corresponds to the desired algorithmic outcome. The following are many examples of PDE techniques applied to medical image analysis.
Stony Brook University Projects

|
Liver Fibrosis Staging by MRI AnalysisIn this project, we provide tools for robust liver fibrosis staging, based on MRI image analysis. The current practice of fibrosis assessment, which is based on painful liver biopsy, might be dangerous. Moreover, the decision of the pathologist based on a biopsy is subjective, and depends on the sample, because the fibrosis level varies along the liver. No objective standard has been developed yet for histological fibrosis assessment. Magnetic resonance volume data has much lower resolution than histological image data, but it includes the entire liver volume. Also, MRI is non-invasive and not painful, thus it is preferred as a diagnostic tool. Previously it has been hypothesized that the average brightness of Apparent Diffusion Coefficient (ADC) in diffusion MRI correlates with the fibrosis stage. More... |
|
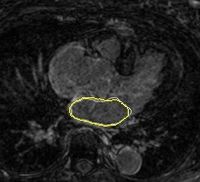
Left Atrium Wall Segmentation Using Topological FeaturesCatheter ablation has been proposed for treatment of atrial fibrillation arrhythmia. MRI data, obtained at University of Utah, are used to explore lesion ablation and scarification locations. In addition, MRI analysis may help to predict if the ablation procedure will help a patient or not. Many of these image analysis tasks are largely based on segmentation of left atrial wall, which is done manually or semi-automatically. Automatic segmentation uses moving contours or surfaces (interfaces) to segment image data by minimizing a predefined energy function. These moving interfaces are highly affected by image data, which can be thought as a force field pushing the interface to features of choice. Thus, the choice of interface attracting image features is critical. More... |
|
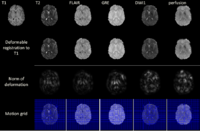
Multimodal Deformable Registration of Traumatic Brain Injury MR VolumesTime-efficient processing and analysis of magnetic resonance imaging (MRI) volumes is desirable for the neurocritical care and monitoring of traumatic brain injury (TBI) patients. An important problem of TBI neuroimaging data analysis is the task of co-registering MR volumes acquired using distinct sequences in the presence of widely variable pixel intensities that are due to the presence of pathology. Here we address this important and challenging problem using an implementation of multimodal deformable registration methods. One method have been implemented on graphics processing units (GPU). In this method we follow a viscous fluid model framework and replace mutual information with the Bhattacharyya distance as the measure of similarity between image volumes. The proposed algorithm is implemented on a GPU and its robustness is illustrated using a longitudinal multimodal TBI dataset. Another method proposes an extension to the principal axis transformation method for finding robust rigid transformation of two volumes. The additional elastic registration is based on a volume registration method MIND, proposed recently by Heinrich et al. More... |
Object Tracking With Adaptive Sobolev Active ContoursIn this project we propose adaptive tracking mechanism which can be used in medical video applications, or 3D volume segmentation. The proposed Sobolev active contour model overcomes the problems of occlusions and changes in scale by adaptive tweaking of the rigidity parameters. The proposed tracking algorithms work in a variety of scenarios and deal naturally with clutter and noise in the scenes, object deformations, partial and entire object occlusions, and low contrast objects. Experimental results show the advantages of our approach compared to state-of-the-art visual trackers.More... | |

|
Multi-scale Shape Representation, Registration, and Segmentation With Applications to RadiotherapyWe present in this work a multiscale representation for shapes with arbitrary topology, and a method to segment the target organ/tissue from medical images having very low contrast with respect to surrounding regions using multiscale shape information and local image features. In a number of previous papers, shape knowledge was incorporated by first constructing a shape space from training data, and then constraining the segmentation process to be within the resulting shape space. However, such an approach has certain limitations including the fact that small scale shape variances may be overwhelmed by those on larger scale, and therefore the local shape information is lost. In this work, first we handle this problem by providing a multiscale shape representation using the wavelet transform. Consequently, the shape variances captured by the statistical learning step are also represented at various scales. In doing so, not only is the diversity of shape enriched, but also small scale changes are nicely captured. More... New: Gao, Y. and Corn, B. and Schifter, D. and Tannenbaum, A. Multiscale 3D Shape Representation and Segmentation with Applications to Hippocampal/Caudate Extraction from Brain MRI, Medical Image Analysis. 16(2) pp374, 2012
|
|
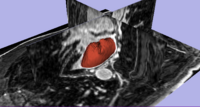
Segmentation and Registration for Atrial Fibrillation Ablation TherapyMagnetic resonance imaging (MRI) has been used for both pre- and and post-ablation assessment of the atrial wall. MRI can aid in selecting the right candidate for the ablation procedure and assessing post-ablation scar formations. Image processing techniques can be used for automatic segmentation of the atrial wall, which facilitates an accurate statistical assessment of the region. As a first step towards the general solution to the computer-assisted segmentation of the left atrial wall, in this research we propose a shape-based image segmentation framework to segment the endocardial wall of the left atrium.More... New: Y. Gao, B. Gholami, R. S. MacLeod, J, Blauer, W. M. Haddad, and A. R. Tannenbaum, Segmentation of the Endocardial Wall of the Left Atrium using Local Region-Based Active Contours and Statistical Shape Learning, SPIE Medical Imaging, San Diego, CA, 2010. |
|
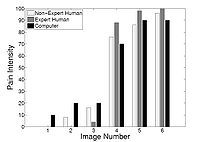
Agitation and Pain Assessment Using Digital ImagingPain assessment in patients who are unable to verbally communicate with medical staff is a challenging problem in patient critical care. The fundamental limitations in sedation and pain assessment in the intensive care unit (ICU) stem from subjective assessment criteria, rather than quantifiable, measurable data for ICU sedation and analgesia. This often results in poor quality and inconsistent treatment of patient agitation and pain from nurse to nurse. Recent advancements in pattern recognition techniques using a relevance vector machine algorithm can assist medical staff in assessing sedation and pain by constantly monitoring the patient and providing the clinician with quantifiable data for ICU sedation. In this paper, we show that the pain intensity assessment given by a computer classifier has a strong correlation with the pain intensity assessed by expert and non-expert human examiners.More... New: B. Gholami, W. M. Haddad, and A. Tannenbaum, Relevance Vector Machine Learning for Neonate Pain Intensity Assessment Using Digital Imaging. IEEE Trans. Biomed. Eng., vol. 57, pp. 1457-1466, 2012. New: B. Gholami, W. M. Haddad, and A. R. Tannenbaum, Agitation and Pain Assessment Using Digital Imaging. Proc. IEEE Eng. Med. Biolog. Conf., Minneapolis, MN, pp. 2176-2179, 2009 (Awarded National Institute of Biomedical Imaging and Bioengineering/National Institute of Health Student Travel Fellowship). New: Wassim M. Haddad, James M. Bailey, Behnood Gholami, and Allen Tannenbaum. Optimal Drug Dosing Control for Intensive Care Unit Sedation Using a Hybrid Deterministic-Stochastic Pharmacokinetic and Pharmacodynamic Model. Optimal Control, Applications and Methods}, 2012, DOI: 10.1002/oca.2038. |
|
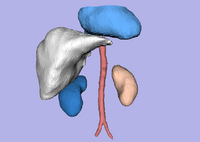
Simultaneous Multiple Object Segmentation using Robust Statistics FeaturesMultiple objects are segmented simultaneously using several interactive active contours based on the feature image which utilizes the robust statistics of the image. More... New: Y. Gao, S. Bouix, M. Shenton, R. Kikinis, A. Tannenbaum. A 3D interactive multi-object segmentation tool using local robust statistics driven active contours. MedIA, volume 16, 2012, pp. 1216-1227. |
|
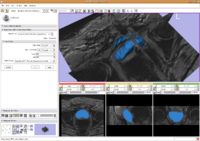
Prostate SegmentationThe 3D prostate MRI images are collected by collaborators at Queen’s University. With a little manual initialization, the algorithm provided the results give to the left. The method mainly uses Random Walk algorithm. More... New: Y. Gao, R. Sandhu, G. Fichtinger, A. Tannenbaum. A coupled global registration and segmentation framework with application to magnetic resonance prostate imagery. IEEE Trans. Medical Imaging, volume 29, 2010, pp. 1781-1794. |

|
Particle Filter Registration of Medical Imagery3D volumetric image registration is performed. The method is based on registering the images through point sets, which is able to handle long distance between as well as registration between Supine and Prone pose prostate. More... New: Y. Gao, R. Sandhu, G. Fichtinger, A. Tannenbaum; A coupled global registration and segmentation framework with application to magnetic resonance prostate imagery. IEEE TMI vol.29, 2010, pp. 1781-1794. New: Y. Gao, Y. Rathi, S. Bouix, A. Tannenbaum; Filtering in the diffeomorphism group and the registration of point sets. IEEE Transactions Image Processing, vol. 21, 2012, pp. 4383-4396 . |
|
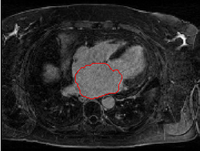
Left Atrium Segmentation for Atrial Fibrillation TreatmentThe planning and evaluation of left atrial ablation procedures is commonly based on the segmentation of the left atrium, which is a challenging task due to large anatomical variations. In this paper, we propose an automatic approach for segmenting the left atrium from magnetic resonance imagery (MRI). The segmentation problem is formulated as a problem in variational region growing. More... New: L. Zhu, Y. Gao, A. Yezzi, A. Tannenbaum. Automatic Segmentation of the Left Atrium from MRI images using Variational Region Growing with a Shape Prior, IEEE Transaction on Image Processing, vol. 22, no. 12, 2013. New: L. Zhu, Y. Gao, A. Yezzi, R. MacLeod, J. Cates, A. Tannenbaum. Automatic Segmentation of the Left Atrium from MRI Images Using Salient Feature and Contour Evolution, IEEE Engineering in Medicine and Biology Conference(EMBC), 2012. |
|
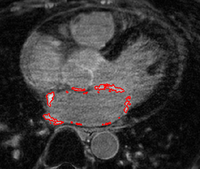
Scar Tissue Identification for Post-Ablation AnalysisThe delay-enhanced MRI (DE-MRI) technique provides an effective way of imaging scarring and fibrosis tissue of atria. Segmentation of the LA from DE-MRI images can be used in atrial fibrillation (AF) treatment to select suitable candidates for ablation therapy and subsequent monitoring of the therapy. More... New: Y. Gao, L. Zhu, A. Yezzi, S. Bouix , A. Tannenbaum. Scar Segmentation in DE-MRI, IEEE International Symposium on Biomedical Imaging (ISBI) , 2012. |

|
Longitudinal Shape Analysis for AFibThe shape evolution of the left atrium in the atrial fibrillation patiens is studied longitudinally to reveal the difference between recover group and the AFib recurrence group. More... |
|
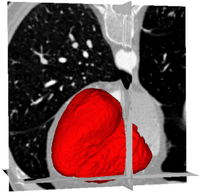
Ventricles Segmentation for Diagnosis of Cardiac DiseasesThis work presents an automatic method for extracting the myocardial wall of the left and right ventricles from cardiac CT images. In the method, the left and right ven- tricles are located sequentially, in which each ventricle is detected by first identifying the endocardial surface and then segmenting the epicardial surface. More... New: L. Zhu, Y. Gao, V. Appia, A. Yezzi, C. Arepalli , A. Stillman, and A. Tannenbaum. Automatic Extraction of the Myocardial Wall from CT Images using Shape Segmentation and Variational Region Growing, IEEE Transaction on Biomedical Engineering, vol. 60, no. 10, 2013. |
|
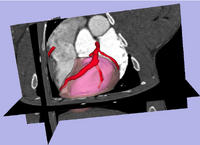
Risk Mass Estimation for Heart Risk EvaluationPrognosis and treatment of cardiovascular diseases frequently require the determination of the myocardial mass at risk caused by coronary stenoses. However, few work has been done for estimating the myocardial mass at risk directly from the heart surface segmented from CAT imagery, rather than using a simplified heart model such as ellipsoid. More... New: L. Zhu, Y. Gao, A. Yezzi, C. Arepalli , A. Stillman, and A. Tannenbaum. A Computational Framework for Estimating the Mass at Risk Caused by Stenoses using CT Angiography, Internatial Journal of Cardiac Imaging(IJCI), In preparation. New: L. Zhu, Y. Gao, V. Mohan, A. Stillman, T. Faber, A. Tannenbaum. Estimation of myocardial volume at risk from CT angiography, Proceedings of SPIE , pp.79632-38A, 2011. |

|
Re-Orientation Approach for Segmentation of DW-MRIThis work proposes a methodology to segment tubular fiber bundles from diffusion weighted magnetic resonance images (DW-MRI). Segmentation is simplified by locally reorienting diffusion information based on large-scale fiber bundle geometry. More... New: Near-Tubular Fiber Bundle Segmentation for Diffusion Weighted Imaging: Segmentation Through Frame Reorientation. Neuroimage, volume 45, 2009, pp. 123-132. |

|
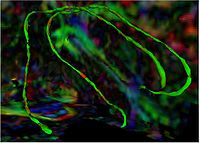
Tubular Surface Segmentation FrameworkWe have proposed a new model for tubular surfaces that transforms the problem of detecting a surface in 3D space, to detecting a curve in 4D space. Besides allowing us to impose a "soft" tubular shape prior, this also leads to computational efficiency over conventional surface segmentation approaches. More... New: V. Mohan, G. Sundaramoorthi, A. Stillman and A. Tannenbaum. Vessel Segmentation with Automatic Centerline Extraction using Tubular Surface Segmentation. September 2009. Proceedings of the Workshop on Cardiac Interventional Imaging and Biophysical Modelling (CI2BM'09), Int Conf Med Image Comput Comput Assist Interv. 2009. New: V. Mohan, G. Sundaramoorthi and A. Tannenbaum. Tubular Surface Segmentation for Extracting Anatomical Structures from Medical Imagery, IEEE Transactions on Medical Imaging, volume 29, 2011, pp. 1945-1958.
|
|
Group Study on DW-MRI using the Tubular Surface ModelWe have proposed a new framework for performing group studies on DW-MRI data sets using the Tubular Surface Model of Mohan et al. We successfully apply this framework to discriminating schizophrenic cases from normal controls, as well as towards visualizing the regions of the Cingulum Bundle that are affected by Schizophrenia. More...
New: V. Mohan, G. Sundaramoorthi, M. Kubicki and A. Tannenbaum. Population Analysis of neural fiber bundles towards schizophrenia detection and characterization, using the Tubular Surface model. Neuroimage (in submission)
|
|
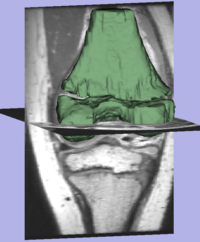
Interactive Image Segmentation With Active ContoursAn approach for tightly coupling the user into a semi-automatic segmentation framework is proposed in this work. A human guides the automatic segmentation by iteratively providing input until convergence to the desired segmentation. The result is a segmentation of manual quality in a fraction of the time; the whole process is intuitive and highly flexible More... New: I. Kolesov, P.Karasev, G.Muller, K.Chudy, J.Xerogeanes, and A. Tannenbaum. Human Supervisory Control Framework for Interactive Medical Image Segmentation. MICCAI Workshop on Computational Biomechanics for Medicine 2011.
|
|
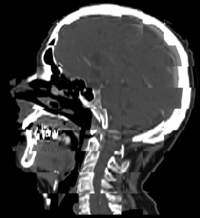
Constrained Registration for Adaptive RadiotherapyA hierarchical approach is described to register two CT scans from different patients. The registration process extracts point clouds representing anatomical structures and aligns them sequentially. The proposed method for registering point clouds can incorporate a variety of constraints including restriction on the injectivity of the deformation field and stationarity of selected landmarks. More... New: I. Kolesov, J. Lee, P.Vela, G. Sharp and A. Tannenbaum. Diffeomorphic Point Set Registration with Landmark Constraints. In Preparation for PAMI.
|
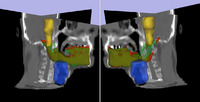
|
Adaptive Radiotherapy for Head, Neck and ThoraxWe proposed an algorithm to include prior knowledge in previously segmented anatomical structures to help in the segmentation of the next structure. This will add enough prior information to allow the Graph Cuts algorithm to segment structures with fuzzy boundaries. More... New: I. Kolesov, V. Mohan, G. Sharp and A. Tannenbaum. Coupled Segmentation for Anatomical Structures by Combining Shape and Relational Spatial Information. MTNS 2010.
|
Kernel Methods for SegmentationSegmentation performances using active contours can be drastically improved if the possible shapes of the object of interest are learnt. The goal of this work is to use Kernel PCA to learn shape priors. Kernel PCA allows for learning nonlinear dependencies in data sets, leading to more robust shape priors. More... New: R. Sandhu, S. Dambreville, and A. Tannenbaum. A Non-Rigid Kernel Based Framework for 2D/3D Pose Estimation and 2D Image Segmentation. IEEE Trans Pattern Anal Mach Intelligence, volume 33, 2011, pp. 1098-1115. | |
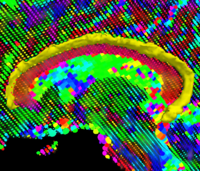
|
Geodesic Tractography SegmentationIn this work, we provide an energy minimization framework which allows one to find fiber tracts and volumetric fiber bundles in brain diffusion-weighted MRI (DW-MRI). More... New: J. Melonakos, E. Pichon, S. Angenent, and A. Tannenbaum. Finsler Active Contours. IEEE Transactions on Pattern Analysis and Machine Intelligence, volume 30, 2008, pp. 412-423. |

|
Label Space: A Coupled Multi-Shape RepresentationMany techniques for multi-shape representation may often develop inaccuracies stemming from either approximations or inherent variation. Label space is an implicit representation that offers unbiased algebraic manipulation and natural expression of label uncertainty. We demonstrate smoothing and registration on multi-label brain MRI. More... New: J. Malcolm, Y. Rathi, S. Bouix, M. Shenton, A. Tannenbaum. Affine registration of label maps in label space. Journal of Computing, volume 2, 2010, pp, 1-11. |
|
Non Parametric Clustering for Biomolecular Structural AnalysisHigh accuracy imaging and image processing techniques allow for collecting structural information of biomolecules with atomistic accuracy. Direct interpretation of the dynamics and the functionality of these structures with physical models, is yet to be developed. Clustering of molecular conformations into classes seems to be the first stage in recovering the formation and the functionality of these molecules. More... New: X. LeFaucheur, E. Hershkovits, R. Tannenbaum, and A. Tannenbaum. Non-parametric clustering for studying RNA conformations. IEEE Trans. Computational Biology and Bioinformatics, volume 8, 2011, pp. 1604-1618. New: Il Tae Kim, A. Tannenbaum, R. Tannenbaum. Anisotropic conductivity of magnetic carbon nanotubes embedded in epoxy matrices. Carbon, volume 49, 2011, pp. 54-61. |
|
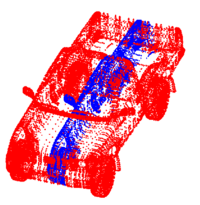
Point Set Rigid RegistrationIn this work, we propose a particle filtering approach for the problem of registering two point sets that differ by a rigid body transformation. Experimental results are provided that demonstrate the robustness of the algorithm to initialization, noise, missing structures or differing point densities in each sets, on challenging 2D and 3D registration tasks. More... New: R. Sandhu, S. Dambreville, A. Tannenbaum. Point set registration via particle filtering and stochastic dynamics. IEEE TPAMI, volume 32, 2010, pp. 1459-1473. New: R. Sandhu, S. Dambreville, A. Tannenbaum. A non-rigid kernel based framework for 2D/3D pose estimation and 2D image segmentation. IEEE TPAMI, volume 33, 2011, pp. 1098-1115. |
|
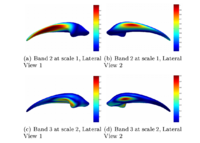
Optimal Mass Transport Registration and VisualizationThe goal of this project is to implement a computationally efficient Elastic/Non-rigid Registration algorithm based on the Monge-Kantorovich theory of optimal mass transport for 3D Medical Imagery. Our technique is based on Multigrid and Multiresolution techniques. This method is particularly useful because it is parameter free and utilizes all of the grayscale data in the image pairs in a symmetric fashion and no landmarks need to be specified for correspondence. More... New: Eldad Haber, Tauseef Rehman, and Allen Tannenbaum. An Efficient Numerical Method for the Solution of the L2 Optimal Mass Transfer Problem. SIAM Journal of Scientific Computing, volume 32, 2011, pp. 197-211. New: Tauseef Rehman, Eldad Haber, Gallagher Pryor, and Allen Tannenbaum. 3D nonrigid registration via optimal mass transport on the GPU. MedIA, volume 13, 2010, pp. 931-940. New: Ayelet Dominitz and Allen Tannenbaum. Texture Mapping Via Optimal Mass Transport. IEEE TVCG, volume 16, 2010), pp. 419-433. |
|
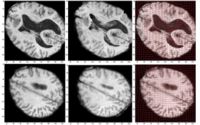
Multiscale Shape Segmentation TechniquesTo represent multiscale variations in a shape population in order to drive the segmentation of deep brain structures, such as the caudate nucleus or the hippocampus. More... |
|
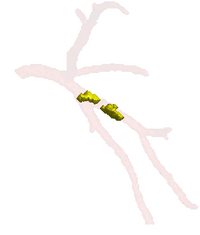
Soft Plaque Detection in CTA ImageryThe ability to detect and measure non-calcified plaques (also known as soft plaques) may improve physicians’ ability to predict cardiac events. This work automatically detects soft plaques in CTA imagery using active contours driven by spatially localized probabilistic models. Plaques are identified by simultaneously segmenting the vessel from the inside-out and the outside-in using carefully chosen localized energies More... New: Soft Plaque Detection and Automatic Vessel Segmentation. PMMIA Workshop in MICCAI, Sep. 2009. |

|
Wavelet Shrinkage for Shape AnalysisShape analysis has become a topic of interest in medical imaging since local variations of a shape could carry relevant information about a disease that may affect only a portion of an organ. We developed a novel wavelet-based denoising and compression statistical model for 3D shapes. More... |
|
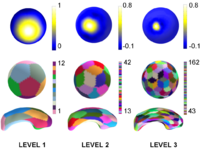
Multiscale Shape AnalysisWe present a novel method of statistical surface-based morphometry based on the use of non-parametric permutation tests and a spherical wavelet (SWC) shape representation. More... |
|
Rule-Based DLPFC SegmentationIn this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the dorsolateral prefrontal cortex. More... |
|
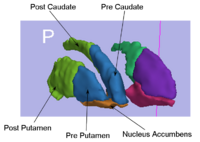
Rule-Based Striatum SegmentationIn this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the striatum. More... |

|
Conformal Surface FlatteningThe goal of this project is for better visualizing and computation of neural activity from fMRI brain imagery. Also, with this technique, shapes can be mapped to shperes for shape analysis, registration or other purposes. Our technique is based on conformal mappings which map genus-zero surface: in fmri case cortical or other surfaces, onto a sphere in an angle preserving manner. More... |
|
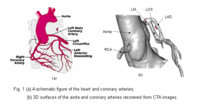
Blood Vessel SegmentationThe goal of this work is to develop blood vessel segmentation techniques for 3D MRI and CT data. The methods have been applied to coronary arteries and portal veins, with promising results. More... New: V.Mohan, G. Sundaramoorthi, A. Stillman and A. Tannenbaum. Vessel Segmentation with Automatic Centerline Extraction Using Tubular Tree Segmentation. CI2BM at MICCAI 2009, September 2009. |
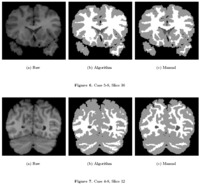
|
Knowledge-Based Bayesian SegmentationThis ITK filter is a segmentation algorithm that utilizes Bayes's Rule along with an affine-invariant anisotropic smoothing filter. More... |
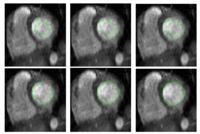
|
Stochastic Methods for SegmentationNew stochastic methods for implementing curvature driven flows for various medical tasks such as segmentation. More... |
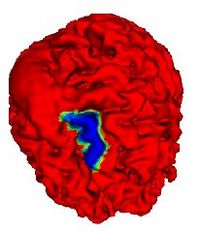
|
Automatic Outlining of sulci on the brain surfaceWe present a method to automatically extract certain key features on a surface. We apply this technique to outline sulci on the cortical surface of a brain. More... |
KPCA, LLE, KLLE Shape AnalysisThe goal of this work is to study and compare shape learning techniques. The techniques considered are Linear Principal Components Analysis (PCA), Kernel PCA, Locally Linear Embedding (LLE) and Kernel LLE. More... | |
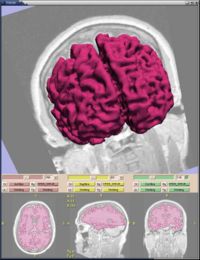
|
Statistical/PDE Methods using Fast Marching for SegmentationThis Fast Marching based flow was added to Slicer 2. More... |