Difference between revisions of "Projects:RegistrationLibrary:RegLib C05"
From NAMIC Wiki
| Line 29: | Line 29: | ||
*[[Image:Button_red_fixed_white.jpg|20px]]reference/fixed : T1 SPGR , 0.9375 x 0.9375 x 1.4 mm voxel size, axial, RAS orientation. | *[[Image:Button_red_fixed_white.jpg|20px]]reference/fixed : T1 SPGR , 0.9375 x 0.9375 x 1.4 mm voxel size, axial, RAS orientation. | ||
*[[Image:Button_green_moving_white.jpg|20px]] moving: T1 SPGR , 0.9375 x 0.9375 x 1.2 mm voxel size, sagittal, RAS orientation. | *[[Image:Button_green_moving_white.jpg|20px]] moving: T1 SPGR , 0.9375 x 0.9375 x 1.2 mm voxel size, sagittal, RAS orientation. | ||
| − | |||
| − | |||
=== Registration Results=== | === Registration Results=== | ||
| Line 52: | Line 50: | ||
=== Discussion: Key Strategies === | === Discussion: Key Strategies === | ||
*the two images have identical contrast, hence we consider "sharper" cost functions, such as NormCorr or MeanSqrd | *the two images have identical contrast, hence we consider "sharper" cost functions, such as NormCorr or MeanSqrd | ||
| − | * | + | *we have aliasing at the image margins that should be masked out |
| − | + | *the two images are not too far apart initially | |
| − | + | *the bone appears largely as signal void, making it hard to distinguish from background | |
| − | * | ||
*because accuracy is more important than speed here, we increase the sampling rate from the default 2% to 15%. | *because accuracy is more important than speed here, we increase the sampling rate from the default 2% to 15%. | ||
*we also expect minimal differences in scale & distortion: so we can either set the expected values to 0 or run a rigid registration | *we also expect minimal differences in scale & distortion: so we can either set the expected values to 0 or run a rigid registration | ||
*we test the result in areas with good anatomical detail and contrast, far away from the pathology. With rigid body motion a local measure of registration accuracy is representative and can give us a valid limit of detectable change. | *we test the result in areas with good anatomical detail and contrast, far away from the pathology. With rigid body motion a local measure of registration accuracy is representative and can give us a valid limit of detectable change. | ||
Revision as of 22:00, 3 February 2010
Home < Projects:RegistrationLibrary:RegLib C05Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
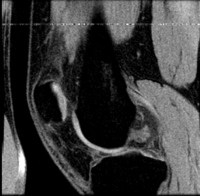
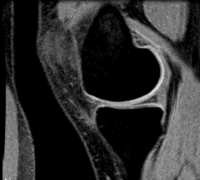
Slicer Registration Use Case Exampe #5: Inter-subject Knee MRI
Objective / Background
The final goal is to align a segmentation prior model to aid in cartilage segmentation.
Keywords
MRI, knee, inter-subject, segmentation
Input Data
 reference/fixed : T1 SPGR , 0.9375 x 0.9375 x 1.4 mm voxel size, axial, RAS orientation.
reference/fixed : T1 SPGR , 0.9375 x 0.9375 x 1.4 mm voxel size, axial, RAS orientation. moving: T1 SPGR , 0.9375 x 0.9375 x 1.2 mm voxel size, sagittal, RAS orientation.
moving: T1 SPGR , 0.9375 x 0.9375 x 1.2 mm voxel size, sagittal, RAS orientation.
Registration Results
Download
Discussion: Registration Challenges
- accuracy is the critical criterion here. We need the registration error (residual misalignment) to be smaller than the change we want to measure/detect. Agreement on what constitutes good alignment can therefore vary greatly.
- the two images have strong differences in coil inhomogeneity. This affects less the registration quality but hampers evaluation. Most of the difference does not become apparent until after registration in direct juxtaposition. Bias field correction beforehand is recommended.
- we have slightly different voxel sizes
- if the pathology change is substantial it might affect the registration.
Discussion: Key Strategies
- the two images have identical contrast, hence we consider "sharper" cost functions, such as NormCorr or MeanSqrd
- we have aliasing at the image margins that should be masked out
- the two images are not too far apart initially
- the bone appears largely as signal void, making it hard to distinguish from background
- because accuracy is more important than speed here, we increase the sampling rate from the default 2% to 15%.
- we also expect minimal differences in scale & distortion: so we can either set the expected values to 0 or run a rigid registration
- we test the result in areas with good anatomical detail and contrast, far away from the pathology. With rigid body motion a local measure of registration accuracy is representative and can give us a valid limit of detectable change.