Difference between revisions of "NA-MIC Collaborations Old"
(New page: <big><big>Resources for New External Collaborators:</big></big> <big><big>Click here</big></big> = NA-MIC External Collaborations = This section describes exte...) |
|||
| Line 263: | Line 263: | ||
# [[NA-MIC/Projects/Diffusion_Image_Analysis/FreeSurfer_NRRD_IO|FreeSurfer NRRD IO]] | # [[NA-MIC/Projects/Diffusion_Image_Analysis/FreeSurfer_NRRD_IO|FreeSurfer NRRD IO]] | ||
# [[Algorithm:MGH:FreeSurferNumericalRecipiesReplacement|FreeSurfer Numerical Recipies Replacement]] | # [[Algorithm:MGH:FreeSurferNumericalRecipiesReplacement|FreeSurfer Numerical Recipies Replacement]] | ||
| − | # [ | + | # [http://www.slicer.org/slicerWiki/index.php/Slicer3:Fluorescence_and_Electron_Microscopy_Support Slicer3 Fluorescence and Electron Microscopy Support] |
Revision as of 21:01, 6 November 2008
Home < NA-MIC Collaborations OldResources for New External Collaborators: Click here
Contents
- 1 NA-MIC External Collaborations
- 1.1 PAR-05-063: R01CA124377 An Integrated System for Image-Guided Radiofrequency Ablation of Liver Tumors
- 1.2 PAR-05-063: R01EB005973 Automated FE Mesh Development
- 1.3 PAR-05-063: R01AA016748 Measuring Alcohol and Stress Interaction with Structural and Perfusion MRI
- 1.4 PAR-05-057: BRAINS Morphology and Image Analysis
- 1.5 Children's Pediatric Cardiology Collaboration with SCI/SPL/Northeastern
- 2 NA-MIC Internal Collaborations
NA-MIC External Collaborations
This section describes external collaborations with NA-MIC that are funded by NIH under the "Collaboration with NCBC" PAR. (Details for this funding mechanism are provided here).
PAR-05-063: R01CA124377 An Integrated System for Image-Guided Radiofrequency Ablation of Liver Tumors
This project is a NCBC collaboration grant. Collaborators include Kevin Cleary and Noby Hata. The master page for this collaboration is here, while the rest of this section contains links to the specific active projects in this collaboration.
Projects
To be added.
PAR-05-063: R01EB005973 Automated FE Mesh Development
This project is a NCBC collaboration grant. Collaborators include Nicole Grosland, Vincent Magnotta, Steve Pieper, and Simon Warfield. The master page for this collaboration is here, while the rest of this section contains links to the specific active projects in this collaboration.
Meshing Algorithms
- Voxel Meshing Module (Iowa)
- Novel Hexahedral Meshing Algorithms (Iowa)
- Hex vs Tet Mesh Comparisons (Iowa/Isomics/BWH)
Automated Segmentation
Image Registration
- Evaluation_of_Inter-Modality_Registration (Iowa/Isomics)
- Inter-slice Motion Correction for fMRI (OSU)
Validation
Mesh Quality Visualization
- FE Mesh Quality Visualization (Iowa/Isomics)
- Standalone FE Mesh Quality Viewer
- Mesh Quality Command Line Module
PAR-05-063: R01AA016748 Measuring Alcohol and Stress Interaction with Structural and Perfusion MRI
This project is a NCBC collaboration grant. Collaborators include James B Daunais, Robert Kraft, Chris Wyatt, William Wells, and Kilian Pohl. The master page for this collaboration is here, while the rest of this section contains links to the specific active projects in this collaboration.
Rhesus Probabalistic Atlas
Segmentation
PAR-05-057: BRAINS Morphology and Image Analysis
This project is not an NCBC collaboration grant, but instead a Continued Development and Maintenance of Software grant. The intent of this application is to update the BRAINS image analysis software developed at the University of Iowa. The collaborators include Vincent Magnotta, Hans Johnson, Jeremy Bockholt, and Nancy Andreasen. There are three main thrusts of this application as outlined below.
Implement a Automated Brain Analysis Pipeline
Code Refactoring for Cross Platform Support
- Refactor Current Modules using ITK and VTK
- Build GUI using Slicer3 Interface
- Use Python as Scripting Language
Validation of Pipeline Using MIND MCIC Sample
- Evaluate MIND MCIC Human Phantom Sample Using Automated Pipeline
- Evaluate Differences between Patients with Schizophrenia and Normal Controls using Automated Pipeline
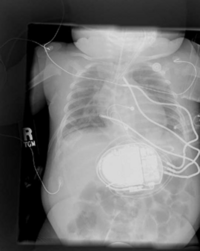
Children's Pediatric Cardiology Collaboration with SCI/SPL/Northeastern
|
Introduction
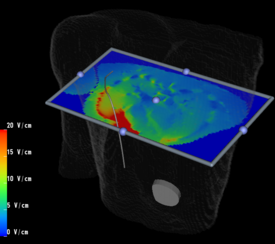
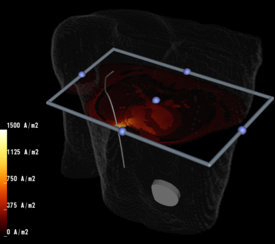
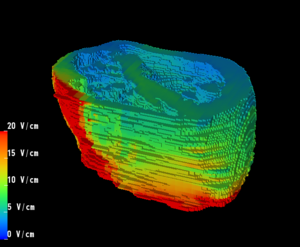
Placement of Implantable Cardiac Defibrillators(ICDs)is a unique and challenging problem for children due to the variety of shapes and sizes, ranging from neonate to adolescent. As a result, a variety of novel implant techniques have been employed. Although these have generally been successful inasmuch as they result in a clinically acceptable defibrillation threshold, nothing is known about the mechanisms by which this threshold is attained, the optimal geometries for defibrillation, and whether unsafe electric field strengths are a result of novel implant approaches. Finite element modeling has been shown in adult torso models to correlate well with clinical results. Our goal is to model defibrillation in child torso models to gain insight into this important problem. We are also interested in developing new orientations in adults with the goal of lowering DFTs and providing options in patients with contraindications to standard techniques.
Goals of Project
1. Create 3D models of children based on CT and MRI datasets for modeling internal and external defibrillation in the SCIRun environment. The processes required address the larger question of how to take any CT or MRI DICOM dataset, segment it into various label maps, combine those label maps in a hierarchical manner, then import and utilize them in the SCIRun/BioPSE environment. This represents part of an expanding collaboration between SCI and SPL to integrate open source tools to allow creation, visualization, and computational modeling of image based 3D models.
2. Create modules which allow insertion of electrode shapes into finite element models in in the SCIRun environment.
3. Utilize the above innovations to model the placement of internal defibrillator electrodes to maximize efficacy, minimize potential cardiac damage, and gain further insight into optimizing defibrillation in children of various sizes as shown above.
Slicer and SPL Tools in the Project
Outline of Torso Segmentation Problem
Segmentation Tools Currently Utilized
Tools In Development and Wishlist
Building and Running SCIRun on SPL Machines
NA-MIC Internal Collaborations
This is a list of all collaborative projects within NA-MIC. These projects form the basis of the progress reports submitted to the NIH. These collaborations are between team members of the various cores of NA-MIC.
Diffusion Image Analysis
Fiber Tract Extraction and Analysis
Continued from 2006
- Fiber Tract Statistics (Utah, UNC)
- Diffusion measures alongs fiber tracts of the cingulum bundle (BWH, MIT, UNC)
- Clustering of anatomically distinct fiber tracts (BWH, MIT)
- Geodesic Active Contours for Fiber Tractography and Fiber Bundle Segmentation (Georgia Tech, BWH)
- Corpus Callosum Fiber Tractography in Schizophrenia (Dartmouth, BWH, MIT)
New in 2007
- Tensor estimation and Monte-Carlo simulation (UNC, Utah)
- ITK Stochastic Tractography Filter (MIT, BWH)
- Volumetric White Matter Connectivity (Utah)
- DTI Atlas-Building (UNC, BWH)
- Inferior Frontotemporal Connections in Schizophrenia (BWH, MIT)
- Automatic Clustering of Corpus Callosum and FA Analysis (BWH, MIT)
- FA Analysis of Fornix Tractography in Schizophrenia (BWH, MIT)
- A Diffusion Tensor Imaging and Genetics Study of Psychosis Across the Lifespan - Project in Recruitment Phase (Toronto, BWH)
Fractional Anisotropy Analysis
Continued from 2006
- Corpus Callosum Regional FA analysis in Schizophrenia (BWH, UNC, Dartmouth)
- Fractional Anisotropy of the Corpus Callosum and Anterior Commissure (BWH, MIT, Dartmouth)
- Fractional Anisotrophy in the Uncinate Fasciculus in Schizophrenia and Bipolar I Disorder--replication and extension of Kubicki study (Dartmouth, BWH)
New in 2007
- Geodesic Active Contours for Fiber Tractography and Fiber Bundle Segmentation (Georgia Tech, BWH)
- Corpus Callosum Probabilistic Subdivision and FA Analysis (BWH, UNC)
Path of Interest Analysis
Continued from 2006
- Integrity of Fronto-Temporal Circuitry in Schizophrenia using Path of Interest Analysis (Dartmouth, MGH, Isomics, BWH)
New in 2007
- ITK implementation of POIStats, and Integration into Slicer3 (MGH, Isomics),
- Dartmouth Data Image Format Compatibility in Application of POIStats (MGH, Isomics, Kitware, Dartmouth)
- Anatomical Connectivity of Regions of Functional Activation (BWH)
Validation
Continued from 2006
- DTI Validation (UCI, MGH, UNC, MIT)
Algorithm/Software Infrastructure
Continued from 2006
- DTI Software/Algorithm Infrastructure (Utah, UNC)
- Tensor based statistics (BWH, Utah)
- Diffusion tensor image filtering (Utah, BWH)
- Non-rigid EPI Registration (MGH, Kitware, Dartmouth)
- Fiber Tools Integration with Slicer 3 (UNC, GE, Isomics)
New in 2007
- Rician Noise Remvoal in Diffusion Tensor MRI (Utah)
- Finsler Tractography in ITK (Georgia Tech, Kitware): 4-block PPT Jan 2007
- Finsler Levelsets in ITK (Georgia Tech, Kitware): 4-block PPT Jan 2007
- Restoration of DWI data using a Rician LMMSE Estimator (BWH)
Structural Image Analysis
Image Segmentation
Continued from 2006
- Knowledge-Based Bayesian Classification and Segmentation (Georgia Tech, Kitware)
- Brain Tissue Classification and Subparcellation of Brain Structures (BWH, MIT, Kitware)
- Rule based segmentation: Striatum (Georgia Tech, BWH, Isomics, Kitware)
- Rule based segmentation: DLPFC (Georgia Tech, UCI, Isomics, Kitware)
- Multiscale Shape Segmentation Techniques (Georgia Tech, BWH)
- Stochastic Methods for Segmentation (Georgia Tech)
- Statistical/PDE methods for Segmentation (Georgia Tech)
- Atlas Renormalization for Improved Brain MR Image Segmentation across Scanner Platforms (MGH)
New in 2007
- EMSegmenter Software Development (BWH, MIT, Kitware) 4-block PPT Jan 2007
- Expanded neuroanatomy training for algorithm development at GA Tech, to expand the rules set for semiautomated segmmentation/shape analysis of frontal cortex (UCI, Georgia Tech)
Image Registration
Continued from 2006
- Optimal Mass Transport for Registration (Georgia Tech, BWH)
New in 2007
- Parallelization of ITK for deformable registration (Kitware, GE, Utah), 4-block PPT Jan 2007
- Group-wise Registration of Medical Images (MIT, BWH, MGH), 4-block PPT Jan 2007
Morphometric Measures and Shape Analysis
Continued from 2006
- Shape Analysis for the caudate and corpus callosum data (BWH, UNC, Georgia Tech, Dartmouth)
- Shape Analysis of the hippocampus (Dartmouth, UNC, BWH)
- Multiscale Shape Analysis applied to Caudate and Hippocampus (Georgia Tech, BWH, UNC)
- UNC Shape Analysis with LONI pipeline for clinical investigators (UNC, UCLA)
- Population Studies (UNC, GE)
- Multi-site morphometry in Mild Cognitive Impairment (UCI, mBIRN?)
- Multi-site morphometry in Schizophrenia (UCI, FBIRN)
- Automated shape model construction (Utah, BWH)
- Neural substrates of apathy in schizophrenia (Dartmouth, Isomics)
- Cortical Surface Shape Analysis Based on Spherical Wavelets (MGH)
- Geometrically-Accurate Topology-Correction of Cortical Surfaces using Non-Separating Loops (MGH)
New in 2007
- ITK Spherical Wavelet Transform Filter (Georgia Tech,GE, Kitware): 4-block PPT Summer 2006, 4-block PPT Jan 2007
- UNC shape analysis with Spherical Wavelet Features (GaTech, UNC): 4-block PPT Jan 2007
- Integrating KWMeshVisu into Slicer (UNC, Kitware), 4-block PPT Jan 2006, 4-block PPT Jan 2007
- Integrating UNC Shape Analysis into Slicer (UNC, Kitware)
- Correspondence of complex structures using (Curvature + Location) MDL (UNC)
- Qdec - a GUI front-end tool for GLM-based group analysis of Freesurfer-processed subject morphometry data (MGH): Freesurfer wiki page
- Adaptive, Particle-Based Sampling for Shapes and Complexes (Utah, UNC)
- Thickness Slicer3 Module (BWH, Isomics)
- Genus Zero Slicer3 Module (BWH, Isomics)
- Morphometry of Frontal circuitry in patients with schizophrenia with and without co-occurring substance use disorder (Dartmouth, Isomics)
fMRI Analysis
Functional Activation Analysis
Continued from 2006
- Neural Substrates of Working Memory in Schizophrenia: A Parametric 3-Back Study (Dartmouth, Harvard)
- Brain Activation during a Continuous Verbal Encoding and Recognition Task in Schizophrenia (Dartmouth, Harvard)
- Fronto-Temporal Connectivity in Schizophrenia during Semantic Memory (Dartmouth, Harvard)
- Imaging Phenotypes in Schizophrenics and Controls (UCI, Toronto)
- Attentional Circuits in Schizophrenia as revealed by fMRI and PET (UCI)
- Serotonergic / dopaminergic genes and processing of threatening stimuli in panic disorder: an imaging genetics study (Toronto)
Algorithm and Software Infrastructure
Continued from 2006
- fMRI Statistics Software Infrastructure (GE, Isomics, Kitware, MIT)
- Spatial Regularization for fMRI Detection (MIT, Harvard)
- Conformal Flattening for fMRI Visualization (Georgia Tech, Harvard)
NA-MIC Kit
NAMIC Software Process
Continued from 2006
- CMake - NAMIC Kit Building (Kitware)
- CPack - NAMIC Kit Distribution (Kitware)
- Dart 2 and CTest - Software Quality (GE, Kitware)
Software Infrastructure
Continued from 2006
- Slicer 3 (Isomics, GE, Kitware, UCSD, UCLA)
- MRML (Isomics, Kitware, GE)
- Coordinate Systems (GE, Isomics)
- IO Unification (GE, Isomics)
- Execution Model (GE)
- Grid Computing (UCSD, GE, Kitware, Isomics)
- Licenses Unification (Kitware, Isomics)
- Toolkits
Training & Dissemination
Continued from 2006
- Training Material and Workshops for NA-MIC Kit (MGH, BWH)
- Dissemination for NA-MIC (Isomics, Kitware, Harvard)