Difference between revisions of "DBP3:Utah"
| (41 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
Back to [[DBP3:Main|NA-MIC DBPs]] | [[Cores|NA-MIC Cores]] | Back to [[DBP3:Main|NA-MIC DBPs]] | [[Cores|NA-MIC Cores]] | ||
| + | === The CARMA CMR Slicer Extension === | ||
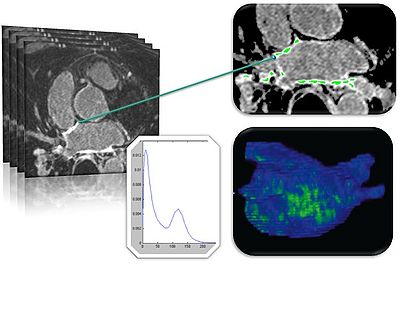
| + | The Slicer CMR Toolkit was developed primarily to support a clinical workflow developed at CARMA and the | ||
| + | University of Utah for (1) segmentation of cardiac structures from MRI, (2) quantification of the amount of fibrosis and scar in the myocardium of cardiac chambers, and (3) visualization of 3D models of cardiac chambers and tissue damage. | ||
| + | |||
| + | For more complete information and to learn how to download the extension, please see | ||
| + | [http://capulet.med.utah.edu/namic/cmrslicer/ the official web page]. | ||
| + | |||
| Line 33: | Line 40: | ||
=== DBP Specific Aims === | === DBP Specific Aims === | ||
| − | + | # Develop and validate image-based longitudinal diagnostic indices for AF | |
| − | + | # Develop automatic segmentation methods for the atrium and adjacent structures | |
| − | + | # Develop an AF scoring scheme to evaluate disease progression and recovery from therapy | |
| − | |||
| − | |||
| − | |||
| − | + | [[Image:TV1.jpg|center|thumb|400px|The CARMA Center's Utah classification for Atrial Fibrillation staging involves segmentation of the left atrial wall from MRI, followed by quantification of enhanced vs. non-enhanced voxels in the wall.]] | |
| + | ==Activities== | ||
| + | === Slicer Modules === | ||
| + | * [[DBP3:Utah:SlicerModuleCMRToolkitWizard|CMR Toolkit Wizard]] | ||
| + | * [[DBP3:Utah:SlicerModuleInhomogeneity|Automatic Cardiac MRI Inhomogeneity Correction Filter]] | ||
| + | * [[DBP3:Utah:SlicerModuleAutoScar|Automatic Left Atrial (Post-ablation) Scar Detection]] | ||
| + | * [[DBP3:Utah:SlicerModuleCardiacRegistration|Cardiac MRI Registration]] | ||
| + | * [[DBP3:Utah:SlicerModuleCardiacRegistrationBRAINSFit|Cardiac MRI Registration Using BRAINSFit]] | ||
| + | * [[DBP3:Utah:SlicerModulePVAntrumCut|PV Antrum Cut Filter]] | ||
| + | * [[DBP3:Utah:SlicerModuleAxialDilate|Axial Dilate Filter]] | ||
| + | * [[DBP3:Utah:SlicerModuleBooleanRemove|Boolean REMOVE Filter]] | ||
| + | * [[DBP3:Utah:SlicerModuleThresholdModel|Threshold Model]] | ||
| + | * [[DBP3:Utah:SlicerModuleIsolatedConnected|Isolated Connected Image Filter]] | ||
| + | * [[DBP3:Utah:SlicerModuleConnectedThreshold|Connected Threshold Image Filter]] | ||
| + | * [[DBP3:Utah:SlicerModuleAutomatedLASegmentation|Automated Graph Cut LA Segmentation]] | ||
| − | === | + | === Tutorials === |
| − | * | + | * [[Media:CARMA_Inhomogeneity_correction_filter_tutorial.pptx|Automatic Cardiac MRI Inhomogeneity Correction Filter Tutorial]] |
| − | ** | + | * [http://wiki.na-mic.org/Wiki/index.php?title=CARMA-LA-Scar_TutorialContestSummer2012 Automatic Left Atrial Scar Tutorial] |
| − | * | + | * [[Media:Carma_Registration_Tutorial.pptx|Cardiac MRI Registration Tutorial]] |
| − | * | + | * [[Media:Carma_BrainsFit_Registration_Tutorial.pptx|Cardiac MRI Registration Using BRAINSFit Tutorial]] |
| + | * [[Media:Carma_PVAntrumCutFilter_Tutorial_New.pptx|PV Antrum Cut Filter Tutorial]] | ||
| + | * [[Media:CARMA_Axial_Dilate_Filter_Tutorial.pptx|Axial Dilate Filter Tutorial]] | ||
| + | * [[Media:Carma_Boolean_Remove_Tutorial.pptx|Boolean REMOVE Filter Tutorial]] | ||
| + | * [[Media:CMRToolkit_Threshold_Model_Tutorial.pptx|Threshold Model Tutorial]] | ||
| + | * [[Media:Carma_Isolated_Connected_Filter_Tutorial.pptx|Isolated Connected Filter Tutorial]] | ||
| + | * [[Media:Carma_Connected_Threshold_Filter_Tutorial.pptx|Connected Threshold Filter Tutorial]] | ||
| + | * [[Media:CarmaWorkflowV2.pdf|Slicer Workflow Tutorial]] | ||
| − | === | + | === Other Projects === |
| − | * | + | * Image registration capabilities for longitudinal analysis: [[DBP3:Utah:RegSegPipeline|Registration & Segmentation pipeline pilots & workflow development (Dominik Meier, Alex Zaitsev)]] |
| − | ** Test and | + | * Evaluation of BRAINSFit and fiducial-based registration tools for LA wall alignment and longitudinal registration of PV: [[DBP3:Utah:PV_BFReg|BRAINSFit Registration of PVs]] |
| + | * Collaboration with GA Tech - | ||
| + | ** Test and evaluate Behnood's wall segmentation - results and analysis are here: [[DBP3:Utah:AutoWallSeg|Automatic Wall Segmentation]] | ||
| + | ** Test and evaluate Yi Gao's atlas-based endocardial segmentation algorithm: [[DBP3:Utah:AutoEndoSeg|Automatic Endocardial Segmentation]] | ||
| + | ** Test and evaluate Yi Gao's image and segmentation-driven registration algorithm: [[DBP3:Utah:VecReg|Image and Segmentation-Driven Registration]] | ||
| − | === | + | === Slicer4 Needs and Plans === |
| − | + | Summary of the Slicer4 needs to support CARMA workflows can be found [[DBP3:Utah:Slicer4 | here]]. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ==Sample Data== | |
| − | *Two | + | * Sixty anonymized datasets containing LGE-MRI image data and segmentations of the left atrium from patients with atrial fibrillation: [http://hdl.handle.net/1926/1755 Pre and Post Radiofrequency Ablation LGE-MRI Collection] |
| + | * Sixty segmentation datasets of the left atrium from patients with atrial fibrillation who have undergone radiofrequency catheter ablation for treatment: [http://hdl.handle.net/1926/1764 CARMA Longitudinal Left Atrial Shape Data] | ||
| + | * Two subjects, two time points: [[Media:CardiacMRIs.zip|CardiacMRIs]]. | ||
Latest revision as of 03:12, 10 January 2014
Home < DBP3:UtahBack to NA-MIC DBPs | NA-MIC Cores
Contents
The CARMA CMR Slicer Extension
The Slicer CMR Toolkit was developed primarily to support a clinical workflow developed at CARMA and the University of Utah for (1) segmentation of cardiac structures from MRI, (2) quantification of the amount of fibrosis and scar in the myocardium of cardiac chambers, and (3) visualization of 3D models of cardiac chambers and tissue damage.
For more complete information and to learn how to download the extension, please see the official web page.
The CARMA DBP: MRI-based study and treatment of atrial fibrillation
Tissue remodeling of the atrial wall is the hallmark of Atrial Fibrillation (AF), a progressive cardiac disease that develops over time (months to years). The mechanisms of of this transformation are only partially understood, but the current scientific focus on tissue remodeling and its putative role in AF suggests an MRI image-based approach to the study of AF.
The Comprehensive Arrhythmia Research and MAnagement (CARMA) Center at the University of Utah is a world leader in the rapidly emerging field of MRI-managed evaluation and ablation of AF. Other groups have begun to recognize the potential of this approach and to investigate and validate some of our findings. Still others are developing new refinements of the MRI technique driven by the specific needs of this application domain.
Preliminary investigation by CARMA has identified image processing and analysis as the rate-limiting step to the development of MRI-based therapies. Novel forms of MRI can be used to evaluate new patients, predict success before ablation, analyze outcomes post-ablation, and guide repeat ablations. Such MRI-based therapies, however, urgently require advanced tools and software to support efficient workflows and accelerate the quantification and analysis of images. The CARMA-NAMIC DBP project will address some of these needs with the development of algorithms and tools for the automated segmentation of heart structures and the MRI-based evaluation of AF progression and recovery.
This remainder of this wiki page describes ongoing CARMA-NAMIC activities and specific plans. For more background information, references, and description of the specific scientific and clinical aims of this project, please see the project web page: http://www.na-mic.org/pages/DBP:Atrial_Fibrillation .
DBP Specific Aims
- Develop and validate image-based longitudinal diagnostic indices for AF
- Develop automatic segmentation methods for the atrium and adjacent structures
- Develop an AF scoring scheme to evaluate disease progression and recovery from therapy
Activities
Slicer Modules
- CMR Toolkit Wizard
- Automatic Cardiac MRI Inhomogeneity Correction Filter
- Automatic Left Atrial (Post-ablation) Scar Detection
- Cardiac MRI Registration
- Cardiac MRI Registration Using BRAINSFit
- PV Antrum Cut Filter
- Axial Dilate Filter
- Boolean REMOVE Filter
- Threshold Model
- Isolated Connected Image Filter
- Connected Threshold Image Filter
- Automated Graph Cut LA Segmentation
Tutorials
- Automatic Cardiac MRI Inhomogeneity Correction Filter Tutorial
- Automatic Left Atrial Scar Tutorial
- Cardiac MRI Registration Tutorial
- Cardiac MRI Registration Using BRAINSFit Tutorial
- PV Antrum Cut Filter Tutorial
- Axial Dilate Filter Tutorial
- Boolean REMOVE Filter Tutorial
- Threshold Model Tutorial
- Isolated Connected Filter Tutorial
- Connected Threshold Filter Tutorial
- Slicer Workflow Tutorial
Other Projects
- Image registration capabilities for longitudinal analysis: Registration & Segmentation pipeline pilots & workflow development (Dominik Meier, Alex Zaitsev)
- Evaluation of BRAINSFit and fiducial-based registration tools for LA wall alignment and longitudinal registration of PV: BRAINSFit Registration of PVs
- Collaboration with GA Tech -
- Test and evaluate Behnood's wall segmentation - results and analysis are here: Automatic Wall Segmentation
- Test and evaluate Yi Gao's atlas-based endocardial segmentation algorithm: Automatic Endocardial Segmentation
- Test and evaluate Yi Gao's image and segmentation-driven registration algorithm: Image and Segmentation-Driven Registration
Slicer4 Needs and Plans
Summary of the Slicer4 needs to support CARMA workflows can be found here.
Sample Data
- Sixty anonymized datasets containing LGE-MRI image data and segmentations of the left atrium from patients with atrial fibrillation: Pre and Post Radiofrequency Ablation LGE-MRI Collection
- Sixty segmentation datasets of the left atrium from patients with atrial fibrillation who have undergone radiofrequency catheter ablation for treatment: CARMA Longitudinal Left Atrial Shape Data
- Two subjects, two time points: CardiacMRIs.